API¶
Workflows¶
fMRIprep base processing workflows¶
-
fmriprep.workflows.base.init_fmriprep_wf(anat_only, aroma_melodic_dim, bold2t1w_dof, cifti_output, debug, dummy_scans, echo_idx, err_on_aroma_warn, fmap_bspline, fmap_demean, force_syn, freesurfer, hires, ignore, layout, longitudinal, low_mem, medial_surface_nan, omp_nthreads, output_dir, output_spaces, regressors_all_comps, regressors_dvars_th, regressors_fd_th, run_uuid, skull_strip_fixed_seed, skull_strip_template, subject_list, t2s_coreg, task_id, use_aroma, use_bbr, use_syn, work_dir)[source]¶ Build fMRIPrep’s pipeline.
This workflow organizes the execution of FMRIPREP, with a sub-workflow for each subject. If FreeSurfer’s
recon-allis to be run, a corresponding folder is created and populated with any needed template subjects under the derivatives folder.- Workflow Graph
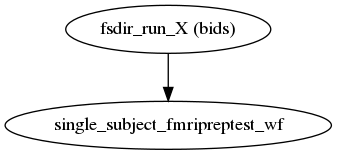
(Source code, png, svg, pdf)
- Parameters
anat_only (bool) – Disable functional workflows
bold2t1w_dof (6, 9 or 12) – Degrees-of-freedom for BOLD-T1w registration
cifti_output (bool) – Generate bold CIFTI file in output spaces
debug (bool) – Enable debugging outputs
dummy_scans (int or None) – Number of volumes to consider as non steady state
echo_idx (int or None) – Index of echo to preprocess in multiecho BOLD series, or
Noneto preprocess allerr_on_aroma_warn (bool) – Do not fail on ICA-AROMA errors
fmap_bspline (bool) – Experimental: Fit B-Spline field using least-squares
fmap_demean (bool) – Demean voxel-shift map during unwarp
force_syn (bool) – Temporary: Always run SyN-based SDC
freesurfer (bool) – Enable FreeSurfer surface reconstruction (may increase runtime)
hires (bool) – Enable sub-millimeter preprocessing in FreeSurfer
ignore (list) – Preprocessing steps to skip (may include “slicetiming”, “fieldmaps”)
layout (BIDSLayout object) – BIDS dataset layout
longitudinal (bool) – Treat multiple sessions as longitudinal (may increase runtime) See sub-workflows for specific differences
low_mem (bool) – Write uncompressed .nii files in some cases to reduce memory usage
medial_surface_nan (bool) – Replace medial wall values with NaNs on functional GIFTI files
omp_nthreads (int) – Maximum number of threads an individual process may use
output_dir (str) – Directory in which to save derivatives
output_spaces (OrderedDict) – Ordered dictionary where keys are TemplateFlow ID strings (e.g.,
MNI152Lin,MNI152NLin6Asym,MNI152NLin2009cAsym, orfsLR) strings designating nonstandard references (e.g.,T1woranat,sbref,run, etc.), or paths pointing to custom templates organized in a TemplateFlow-like structure. Values of the dictionary aggregate modifiers (e.g., the value for the keyMNI152Lincould be{'resolution': 2}if one wants the resampling to be done on the 2mm resolution version of the selected template).regressors_all_comps – Return all CompCor component time series instead of the top fraction
regressors_dvars_th – Criterion for flagging DVARS outliers
regressors_fd_th – Criterion for flagging framewise displacement outliers
run_uuid (str) – Unique identifier for execution instance
skull_strip_template (tuple) – Name of target template for brain extraction with ANTs’
antsBrainExtraction, and corresponding dictionary of output-space modifiers.skull_strip_fixed_seed (bool) – Do not use a random seed for skull-stripping - will ensure run-to-run replicability when used with –omp-nthreads 1
subject_list (list) – List of subject labels
t2s_coreg (bool) – For multi-echo EPI, use the calculated T2*-map for T2*-driven coregistration
task_id (str or None) – Task ID of BOLD series to preprocess, or
Noneto preprocess alluse_aroma (bool) – Perform ICA-AROMA on MNI-resampled functional series
use_bbr (bool or None) – Enable/disable boundary-based registration refinement. If
None, test BBR result for distortion before accepting.use_syn (bool) – Experimental: Enable ANTs SyN-based susceptibility distortion correction (SDC). If fieldmaps are present and enabled, this is not run, by default.
work_dir (str) – Directory in which to store workflow execution state and temporary files
-
fmriprep.workflows.base.init_single_subject_wf(anat_only, aroma_melodic_dim, bold2t1w_dof, cifti_output, debug, dummy_scans, echo_idx, err_on_aroma_warn, fmap_bspline, fmap_demean, force_syn, freesurfer, hires, ignore, layout, longitudinal, low_mem, medial_surface_nan, name, omp_nthreads, output_dir, output_spaces, reportlets_dir, regressors_all_comps, regressors_dvars_th, regressors_fd_th, skull_strip_fixed_seed, skull_strip_template, subject_id, t2s_coreg, task_id, use_aroma, use_bbr, use_syn)[source]¶ This workflow organizes the preprocessing pipeline for a single subject.
It collects and reports information about the subject, and prepares sub-workflows to perform anatomical and functional preprocessing. Anatomical preprocessing is performed in a single workflow, regardless of the number of sessions. Functional preprocessing is performed using a separate workflow for each individual BOLD series.
- Workflow Graph
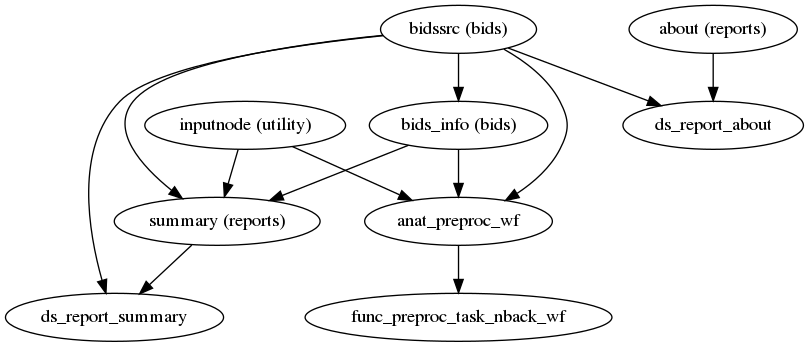
(Source code, png, svg, pdf)
- Parameters
anat_only (bool) – Disable functional workflows
aroma_melodic_dim (int) – Maximum number of components identified by MELODIC within ICA-AROMA (default is -200, i.e., no limitation).
bold2t1w_dof (6, 9 or 12) – Degrees-of-freedom for BOLD-T1w registration
cifti_output (bool) – Generate bold CIFTI file in output spaces
debug (bool) – Enable debugging outputs
dummy_scans (int or None) – Number of volumes to consider as non steady state
echo_idx (int or None) – Index of echo to preprocess in multiecho BOLD series, or
Noneto preprocess allerr_on_aroma_warn (bool) – Do not fail on ICA-AROMA errors
fmap_bspline (bool) – Experimental: Fit B-Spline field using least-squares
fmap_demean (bool) – Demean voxel-shift map during unwarp
force_syn (bool) – Temporary: Always run SyN-based SDC
freesurfer (bool) – Enable FreeSurfer surface reconstruction (may increase runtime)
hires (bool) – Enable sub-millimeter preprocessing in FreeSurfer
ignore (list) – Preprocessing steps to skip (may include “slicetiming”, “fieldmaps”)
layout (BIDSLayout object) – BIDS dataset layout
longitudinal (bool) – Treat multiple sessions as longitudinal (may increase runtime) See sub-workflows for specific differences
low_mem (bool) – Write uncompressed .nii files in some cases to reduce memory usage
medial_surface_nan (bool) – Replace medial wall values with NaNs on functional GIFTI files
name (str) – Name of workflow
omp_nthreads (int) – Maximum number of threads an individual process may use
output_dir (str) – Directory in which to save derivatives
output_spaces (OrderedDict) – Ordered dictionary where keys are TemplateFlow ID strings (e.g.,
MNI152Lin,MNI152NLin6Asym,MNI152NLin2009cAsym, orfsLR) strings designating nonstandard references (e.g.,T1woranat,sbref,run, etc.), or paths pointing to custom templates organized in a TemplateFlow-like structure. Values of the dictionary aggregate modifiers (e.g., the value for the keyMNI152Lincould be{'resolution': 2}if one wants the resampling to be done on the 2mm resolution version of the selected template).reportlets_dir (str) – Directory in which to save reportlets
regressors_all_comps – Return all CompCor component time series instead of the top fraction
regressors_fd_th – Criterion for flagging framewise displacement outliers
regressors_dvars_th – Criterion for flagging DVARS outliers
skull_strip_fixed_seed (bool) – Do not use a random seed for skull-stripping - will ensure run-to-run replicability when used with –omp-nthreads 1
skull_strip_template (tuple) – Name of target template for brain extraction with ANTs’
antsBrainExtraction, and corresponding dictionary of output-space modifiers.subject_id (str) – List of subject labels
t2s_coreg (bool) – For multi-echo EPI, use the calculated T2*-map for T2*-driven coregistration
task_id (str or None) – Task ID of BOLD series to preprocess, or
Noneto preprocess alluse_aroma (bool) – Perform ICA-AROMA on MNI-resampled functional series
use_bbr (bool or None) – Enable/disable boundary-based registration refinement. If
None, test BBR result for distortion before accepting.use_syn (bool) – Experimental: Enable ANTs SyN-based susceptibility distortion correction (SDC). If fieldmaps are present and enabled, this is not run, by default.
- Inputs
subjects_dir (str) – FreeSurfer’s
$SUBJECTS_DIR.
Anatomical reference preprocessing workflows¶
-
fmriprep.workflows.anatomical.init_anat_preproc_wf(bids_root, freesurfer, hires, longitudinal, omp_nthreads, output_dir, output_spaces, num_t1w, reportlets_dir, skull_strip_template, debug=False, name='anat_preproc_wf', skull_strip_fixed_seed=False)[source]¶ Stage the anatomical preprocessing steps of sMRIPrep.
This includes:
T1w reference: realigning and then averaging T1w images.
Brain extraction and INU (bias field) correction.
Brain tissue segmentation.
Spatial normalization to standard spaces.
Surface reconstruction with FreeSurfer.
- Workflow Graph
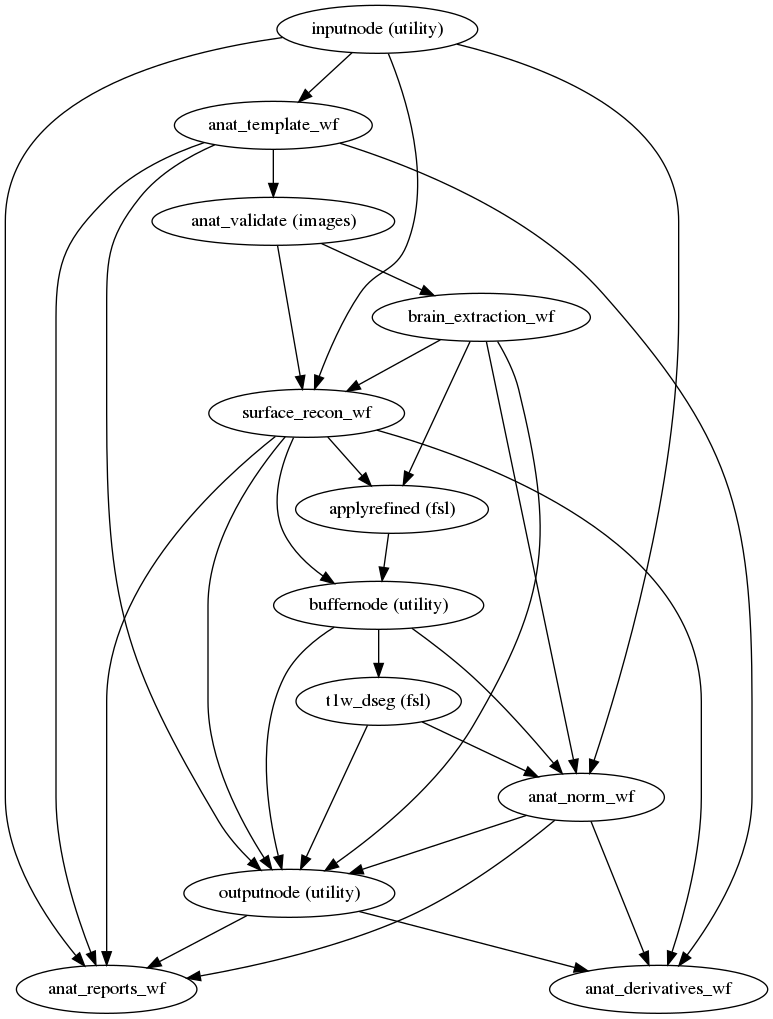
(Source code, png, svg, pdf)
- Subworkflows
init_brain_extraction_wf()init_surface_recon_wf()
- Parameters
bids_root (str) – Path of the input BIDS dataset root
debug (bool) – Enable debugging outputs
freesurfer (bool) – Enable FreeSurfer surface reconstruction (increases runtime by 6h, at the very least)
output_spaces (list) – List of spatial normalization targets. Some parts of pipeline will only be instantiated for some output spaces. Valid spaces:
Any template identifier from TemplateFlow
Path to a template folder organized following TemplateFlow’s conventions
hires (bool) – Enable sub-millimeter preprocessing in FreeSurfer
longitudinal (bool) – Create unbiased structural template, regardless of number of inputs (may increase runtime)
name (str, optional) – Workflow name (default: anat_preproc_wf)
omp_nthreads (int) – Maximum number of threads an individual process may use
output_dir (str) – Directory in which to save derivatives
reportlets_dir (str) – Directory in which to save reportlets
skull_strip_fixed_seed (bool) – Do not use a random seed for skull-stripping - will ensure run-to-run replicability when used with –omp-nthreads 1 (default:
False).skull_strip_template (tuple) – Name of ANTs skull-stripping template and specifications.
- Inputs
t1w – List of T1-weighted structural images
t2w – List of T2-weighted structural images
flair – List of FLAIR images
subjects_dir – FreeSurfer SUBJECTS_DIR
- Outputs
t1w_preproc – The T1w reference map, which is calculated as the average of bias-corrected and preprocessed T1w images, defining the anatomical space.
t1w_brain – Skull-stripped
t1w_preproct1w_mask – Brain (binary) mask estimated by brain extraction.
t1w_dseg – Brain tissue segmentation of the preprocessed structural image, including gray-matter (GM), white-matter (WM) and cerebrospinal fluid (CSF).
t1w_tpms – List of tissue probability maps corresponding to
t1w_dseg.std_t1w – T1w reference resampled in one or more standard spaces.
std_mask – Mask of skull-stripped template, in MNI space
std_dseg – Segmentation, resampled into MNI space
std_tpms – List of tissue probability maps in MNI space
subjects_dir – FreeSurfer SUBJECTS_DIR
anat2std_xfm – Nonlinear spatial transform to resample imaging data given in anatomical space into standard space.
std2anat_xfm – Inverse transform of the above.
subject_id – FreeSurfer subject ID
t1w2fsnative_xfm – LTA-style affine matrix translating from T1w to FreeSurfer-conformed subject space
fsnative2t1w_xfm – LTA-style affine matrix translating from FreeSurfer-conformed subject space to T1w
surfaces – GIFTI surfaces (gray/white boundary, midthickness, pial, inflated)
Surface preprocessing¶
fMRIPrep uses FreeSurfer to reconstruct surfaces from T1w/T2w structural images.
-
fmriprep.workflows.anatomical.init_surface_recon_wf(omp_nthreads, hires, name='surface_recon_wf')[source]¶ Reconstruct anatomical surfaces using FreeSurfer’s
recon-all.Reconstruction is performed in three phases. The first phase initializes the subject with T1w and T2w (if available) structural images and performs basic reconstruction (
autorecon1) with the exception of skull-stripping. For example, a subject with only one session with T1w and T2w images would be processed by the following command:$ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -i <bids-root>/sub-<subject_label>/anat/sub-<subject_label>_T1w.nii.gz \ -T2 <bids-root>/sub-<subject_label>/anat/sub-<subject_label>_T2w.nii.gz \ -autorecon1 \ -noskullstripThe second phase imports an externally computed skull-stripping mask. This workflow refines the external brainmask using the internal mask implicit the the FreeSurfer’s
aseg.mgzsegmentation, to reconcile ANTs’ and FreeSurfer’s brain masks.First, the
aseg.mgzmask from FreeSurfer is refined in two steps, using binary morphological operations:With a binary closing operation the sulci are included into the mask. This results in a smoother brain mask that does not exclude deep, wide sulci.
Fill any holes (typically, there could be a hole next to the pineal gland and the corpora quadrigemina if the great cerebral brain is segmented out).
Second, the brain mask is grown, including pixels that have a high likelihood to the GM tissue distribution:
Dilate and substract the brain mask, defining the region to search for candidate pixels that likely belong to cortical GM.
Pixels found in the search region that are labeled as GM by ANTs (during
antsBrainExtraction.sh) are directly added to the new mask.Otherwise, estimate GM tissue parameters locally in patches of
wwsize, and test the likelihood of the pixel to belong in the GM distribution.
This procedure is inspired on mindboggle’s solution to the problem: https://github.com/nipy/mindboggle/blob/7f91faaa7664d820fe12ccc52ebaf21d679795e2/mindboggle/guts/segment.py#L1660
The final phase resumes reconstruction, using the T2w image to assist in finding the pial surface, if available. See
init_autorecon_resume_wf()for details.Memory annotations for FreeSurfer are based off their documentation. They specify an allocation of 4GB per subject. Here we define 5GB to have a certain margin.
- Workflow Graph
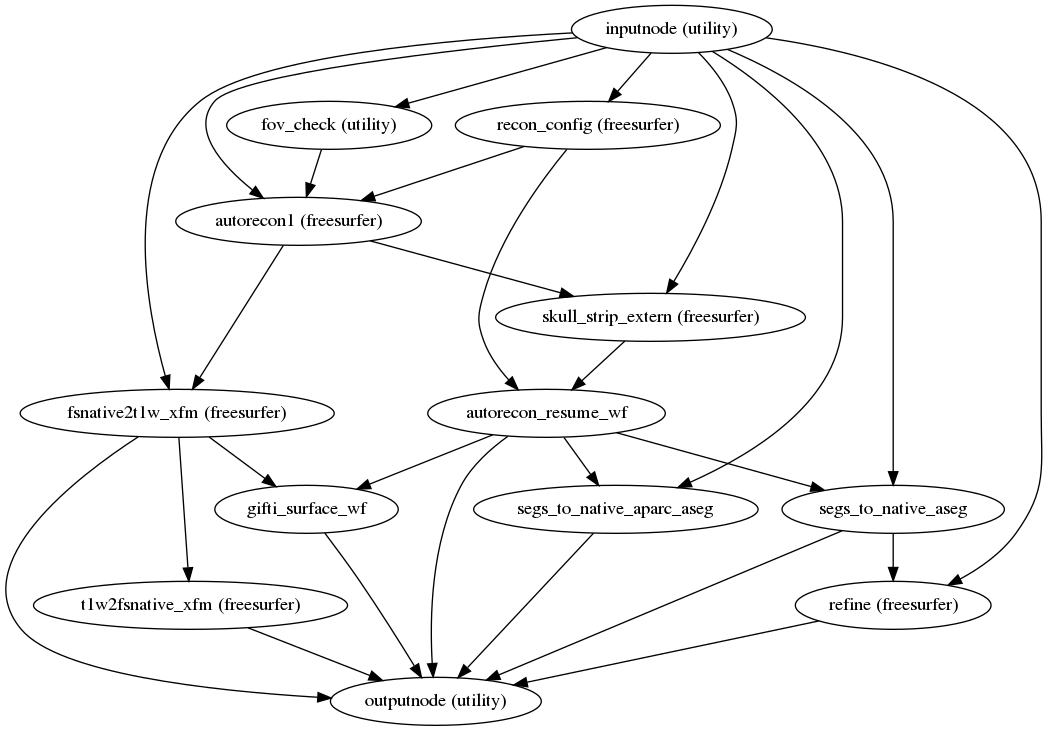
(Source code, png, svg, pdf)
- Subworkflows
init_autorecon_resume_wf()init_gifti_surface_wf()
- Parameters
omp_nthreads (int) – Maximum number of threads an individual process may use
hires (bool) – Enable sub-millimeter preprocessing in FreeSurfer
- Inputs
t1w – List of T1-weighted structural images
t2w – List of T2-weighted structural images (only first used)
flair – List of FLAIR images
skullstripped_t1 – Skull-stripped T1-weighted image (or mask of image)
ants_segs – Brain tissue segmentation from ANTS
antsBrainExtraction.shcorrected_t1 – INU-corrected, merged T1-weighted image
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
- Outputs
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
t1w2fsnative_xfm – LTA-style affine matrix translating from T1w to FreeSurfer-conformed subject space
fsnative2t1w_xfm – LTA-style affine matrix translating from FreeSurfer-conformed subject space to T1w
surfaces – GIFTI surfaces for gray/white matter boundary, pial surface, midthickness (or graymid) surface, and inflated surfaces
out_brainmask – Refined brainmask, derived from FreeSurfer’s
asegvolumeout_aseg – FreeSurfer’s aseg segmentation, in native T1w space
out_aparc – FreeSurfer’s aparc+aseg segmentation, in native T1w space
-
fmriprep.workflows.anatomical.init_autorecon_resume_wf(omp_nthreads, name='autorecon_resume_wf')[source]¶ Resume recon-all execution, assuming the -autorecon1 stage has been completed.
In order to utilize resources efficiently, this is broken down into seven sub-stages; after the first stage, the second and third stages may be run simultaneously, and the fifth and sixth stages may be run simultaneously, if resources permit; the fourth stage must be run prior to the fifth and sixth, and the seventh must be run after:
$ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -autorecon2-volonly $ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -autorecon-hemi lh -T2pial \ -noparcstats -noparcstats2 -noparcstats3 -nohyporelabel -nobalabels $ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -autorecon-hemi rh -T2pial \ -noparcstats -noparcstats2 -noparcstats3 -nohyporelabel -nobalabels $ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -cortribbon $ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -autorecon-hemi lh -nohyporelabel $ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -autorecon-hemi rh -nohyporelabel $ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -autorecon3The parcellation statistics steps are excluded from the second and third stages, because they require calculation of the cortical ribbon volume (the fourth stage). Hypointensity relabeling is excluded from hemisphere-specific steps to avoid race conditions, as it is a volumetric operation.
- Workflow Graph
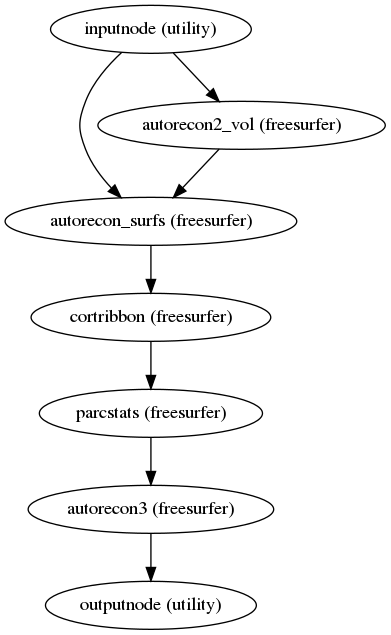
(Source code, png, svg, pdf)
- Inputs
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
use_T2 – Refine pial surface using T2w image
use_FLAIR – Refine pial surface using FLAIR image
- Outputs
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
-
fmriprep.workflows.anatomical.init_gifti_surface_wf(name='gifti_surface_wf')[source]¶ Prepare GIFTI surfaces from a FreeSurfer subjects directory.
If midthickness (or graymid) surfaces do not exist, they are generated and saved to the subject directory as
lh/rh.midthickness. These, along with the gray/white matter boundary (lh/rh.smoothwm), pial sufaces (lh/rh.pial) and inflated surfaces (lh/rh.inflated) are converted to GIFTI files. Additionally, the vertex coordinates arerecenteredto align with native T1w space.- Workflow Graph
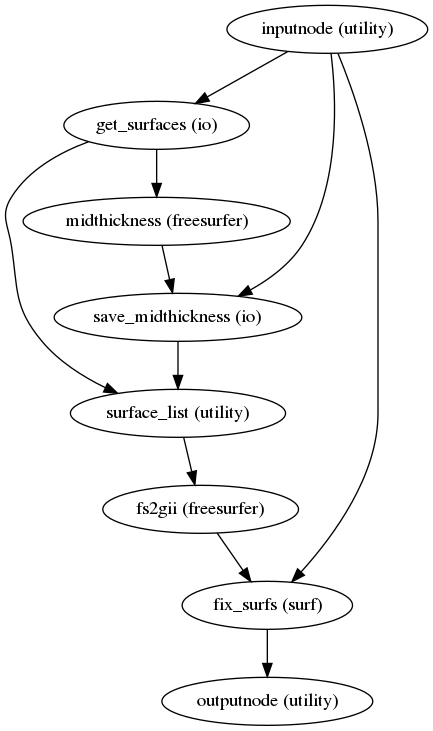
(Source code, png, svg, pdf)
- Inputs
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
fsnative2t1w_xfm – LTA formatted affine transform file (inverse)
- Outputs
surfaces – GIFTI surfaces for gray/white matter boundary, pial surface, midthickness (or graymid) surface, and inflated surfaces
Pre-processing fMRI - BOLD signal workflows¶
Orchestrating the BOLD-preprocessing workflow¶
-
fmriprep.workflows.bold.base.init_func_preproc_wf(aroma_melodic_dim, bold2t1w_dof, bold_file, cifti_output, debug, dummy_scans, err_on_aroma_warn, fmap_bspline, fmap_demean, force_syn, freesurfer, ignore, low_mem, medial_surface_nan, omp_nthreads, output_dir, output_spaces, regressors_all_comps, regressors_dvars_th, regressors_fd_th, reportlets_dir, t2s_coreg, use_aroma, use_bbr, use_syn, layout=None, num_bold=1)[source]¶ This workflow controls the functional preprocessing stages of fMRIPrep.
- Workflow Graph
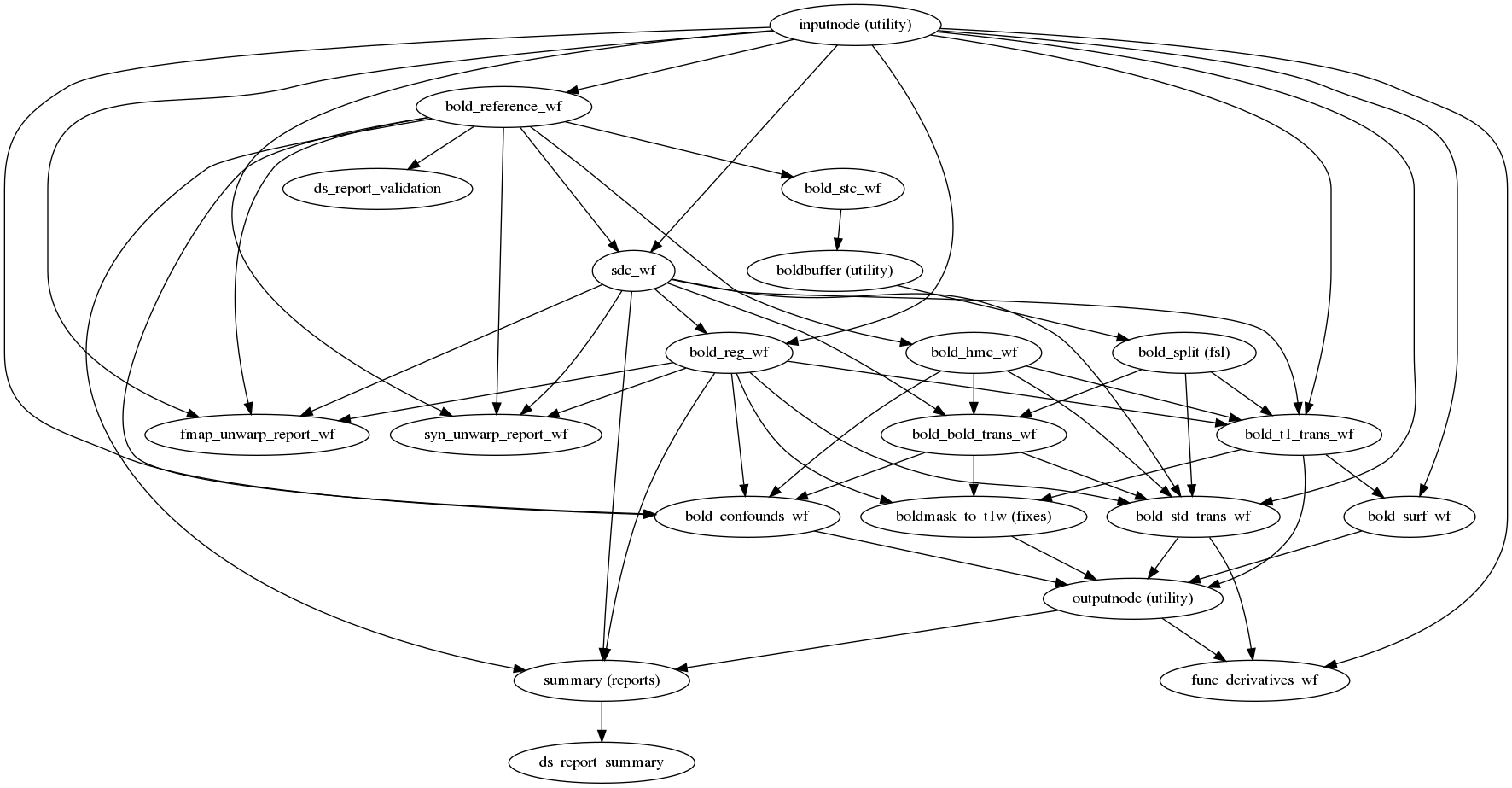
(Source code, png, svg, pdf)
- Parameters
aroma_melodic_dim (int) – Maximum number of components identified by MELODIC within ICA-AROMA (default is -200, ie. no limitation).
bold2t1w_dof (6, 9 or 12) – Degrees-of-freedom for BOLD-T1w registration
bold_file (str) – BOLD series NIfTI file
cifti_output (bool) – Generate bold CIFTI file in output spaces
debug (bool) – Enable debugging outputs
dummy_scans (int or None) – Number of volumes to consider as non steady state
err_on_aroma_warn (bool) – Do not crash on ICA-AROMA errors
fmap_bspline (bool) – Experimental: Fit B-Spline field using least-squares
fmap_demean (bool) – Demean voxel-shift map during unwarp
force_syn (bool) – Temporary: Always run SyN-based SDC
freesurfer (bool) – Enable FreeSurfer functional registration (bbregister) and resampling BOLD series to FreeSurfer surface meshes.
ignore (list) – Preprocessing steps to skip (may include “slicetiming”, “fieldmaps”)
low_mem (bool) – Write uncompressed .nii files in some cases to reduce memory usage
medial_surface_nan (bool) – Replace medial wall values with NaNs on functional GIFTI files
omp_nthreads (int) – Maximum number of threads an individual process may use
output_dir (str) – Directory in which to save derivatives
output_spaces (OrderedDict) – Ordered dictionary where keys are TemplateFlow ID strings (e.g.
MNI152Lin,MNI152NLin6Asym,MNI152NLin2009cAsym, orfsLR) strings designating nonstandard references (e.g.T1woranat,sbref,run, etc.), or paths pointing to custom templates organized in a TemplateFlow-like structure. Values of the dictionary aggregate modifiers (e.g. the value for the keyMNI152Lincould be{'resolution': 2}if one wants the resampling to be done on the 2mm resolution version of the selected template).regressors_all_comps – Return all CompCor component time series instead of the top fraction
regressors_dvars_th – Criterion for flagging DVARS outliers
regressors_fd_th – Criterion for flagging framewise displacement outliers
reportlets_dir (str) – Absolute path of a directory in which reportlets will be temporarily stored
t2s_coreg (bool) – For multiecho EPI, use the calculated T2*-map for T2*-driven coregistration
use_aroma (bool) – Perform ICA-AROMA on MNI-resampled functional series
use_bbr (bool or None) – Enable/disable boundary-based registration refinement. If
None, test BBR result for distortion before accepting. When usingt2s_coreg, BBR will be enabled by default unless explicitly specified otherwise.use_syn (bool) – Experimental: Enable ANTs SyN-based susceptibility distortion correction (SDC). If fieldmaps are present and enabled, this is not run, by default.
layout (BIDSLayout) – BIDSLayout structure to enable metadata retrieval
num_bold (int) – Total number of BOLD files that have been set for preprocessing (default is 1)
- Inputs
bold_file – BOLD series NIfTI file
t1w_preproc – Bias-corrected structural template image
t1w_brain – Skull-stripped
t1w_preproct1w_mask – Mask of the skull-stripped template image
t1w_dseg – Segmentation of preprocessed structural image, including gray-matter (GM), white-matter (WM) and cerebrospinal fluid (CSF)
t1w_asec – Segmentation of structural image, done with FreeSurfer.
t1w_aparc – Parcellation of structural image, done with FreeSurfer.
t1w_tpms – List of tissue probability maps in T1w space
anat2std_xfm – ANTs-compatible affine-and-warp transform file
std2anat_xfm – ANTs-compatible affine-and-warp transform file (inverse)
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
t1w2fsnative_xfm – LTA-style affine matrix translating from T1w to FreeSurfer-conformed subject space
fsnative2t1w_xfm – LTA-style affine matrix translating from FreeSurfer-conformed subject space to T1w
- Outputs
bold_t1 – BOLD series, resampled to T1w space
bold_mask_t1 – BOLD series mask in T1w space
bold_std – BOLD series, resampled to template space
bold_mask_std – BOLD series mask in template space
confounds – TSV of confounds
surfaces – BOLD series, resampled to FreeSurfer surfaces
aroma_noise_ics – Noise components identified by ICA-AROMA
melodic_mix – FSL MELODIC mixing matrix
bold_cifti – BOLD CIFTI image
cifti_variant – combination of target spaces for bold_cifti
-
fmriprep.workflows.bold.base.init_func_derivatives_wf(bids_root, cifti_output, freesurfer, metadata, output_dir, output_spaces, standard_spaces, use_aroma, name='func_derivatives_wf')[source]¶ Set up a battery of datasinks to store derivatives in the right location.
- Parameters
bids_root (str) – Original BIDS dataset path.
cifti_output (bool) – Whether the
--cifti-outputflag was set.freesurfer (bool) – Whether FreeSurfer anatomical processing was run.
metadata (dict) – Metadata dictionary associated to the BOLD run.
output_dir (str) – Where derivatives should be written out to.
output_spaces (OrderedDict) – List of selected
--output-spaces.use_aroma (bool) – Whether
--use-aromaflag was set.name (str) – This workflow’s identifier (default:
func_derivatives_wf).
Utility workflows¶
-
fmriprep.workflows.bold.util.init_bold_reference_wf(omp_nthreads, bold_file=None, pre_mask=False, name='bold_reference_wf', gen_report=False)[source]¶ Build a workflow that generates reference BOLD images for a series.
The raw reference image is the target of HMC, and a contrast-enhanced reference is the subject of distortion correction, as well as boundary-based registration to T1w and template spaces.
- Workflow Graph
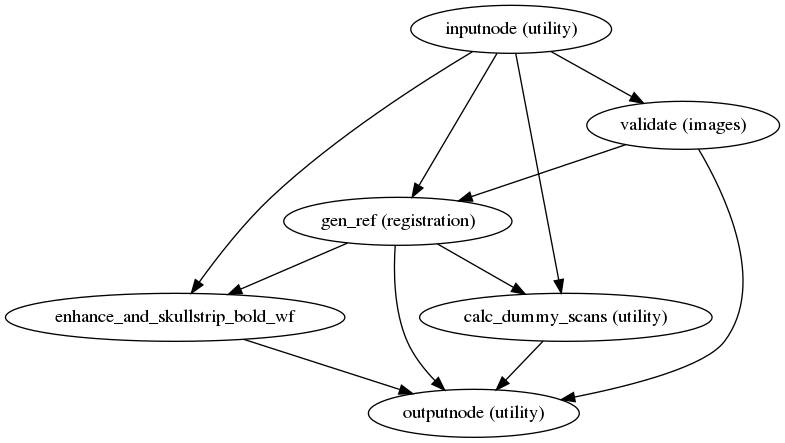
(Source code, png, svg, pdf)
- Parameters
bold_file (str) – BOLD series NIfTI file
omp_nthreads (int) – Maximum number of threads an individual process may use
name (str) – Name of workflow (default:
bold_reference_wf)gen_report (bool) – Whether a mask report node should be appended in the end
- Inputs
bold_file (str) – BOLD series NIfTI file
bold_mask (bool) – A tentative brain mask to initialize the workflow (requires
pre_maskparameter setTrue).dummy_scans (int or None) – Number of non-steady-state volumes specified by user at beginning of
bold_filesbref_file (str) – single band (as opposed to multi band) reference NIfTI file
- Outputs
bold_file (str) – Validated BOLD series NIfTI file
raw_ref_image (str) – Reference image to which BOLD series is motion corrected
skip_vols (int) – Number of non-steady-state volumes selected at beginning of
bold_filealgo_dummy_scans (int) – Number of non-steady-state volumes agorithmically detected at beginning of
bold_fileref_image (str) – Contrast-enhanced reference image
ref_image_brain (str) – Skull-stripped reference image
bold_mask (str) – Skull-stripping mask of reference image
validation_report (str) – HTML reportlet indicating whether
bold_filehad a valid affine
- Subworkflows
init_enhance_and_skullstrip_wf()
-
fmriprep.workflows.bold.util.init_enhance_and_skullstrip_bold_wf(name='enhance_and_skullstrip_bold_wf', pre_mask=False, omp_nthreads=1)[source]¶ Enhance and run brain extraction on a BOLD EPI image.
This workflow takes in a BOLD fMRI average/summary (e.g., a reference image averaging non-steady-state timepoints), and sharpens the histogram with the application of the N4 algorithm for removing the INU bias field and calculates a signal mask.
Steps of this workflow are:
Calculate a tentative mask by registering (9-parameters) to fMRIPrep’s EPI -boldref template, which is in MNI space. The tentative mask is obtained by resampling the MNI template’s brainmask into boldref-space.
Binary dilation of the tentative mask with a sphere of 3mm diameter.
Run ANTs’
N4BiasFieldCorrectionon the input BOLD average, using the mask generated in 1) instead of the internal Otsu thresholding.Calculate a loose mask using FSL’s
bet, with one mathematical morphology dilation of one iteration and a sphere of 6mm as structuring element.Mask the INU-corrected image with the latest mask calculated in 3), then use AFNI’s
3dUnifizeto standardize the T2* contrast distribution.Calculate a mask using AFNI’s
3dAutomaskafter the contrast enhancement of 4).Calculate a final mask as the intersection of 4) and 6).
Apply final mask on the enhanced reference.
Step 1 can be skipped if the
pre_maskargument is set toTrueand a tentative mask is passed in to the workflow throught thepre_maskNipype input.- Workflow graph
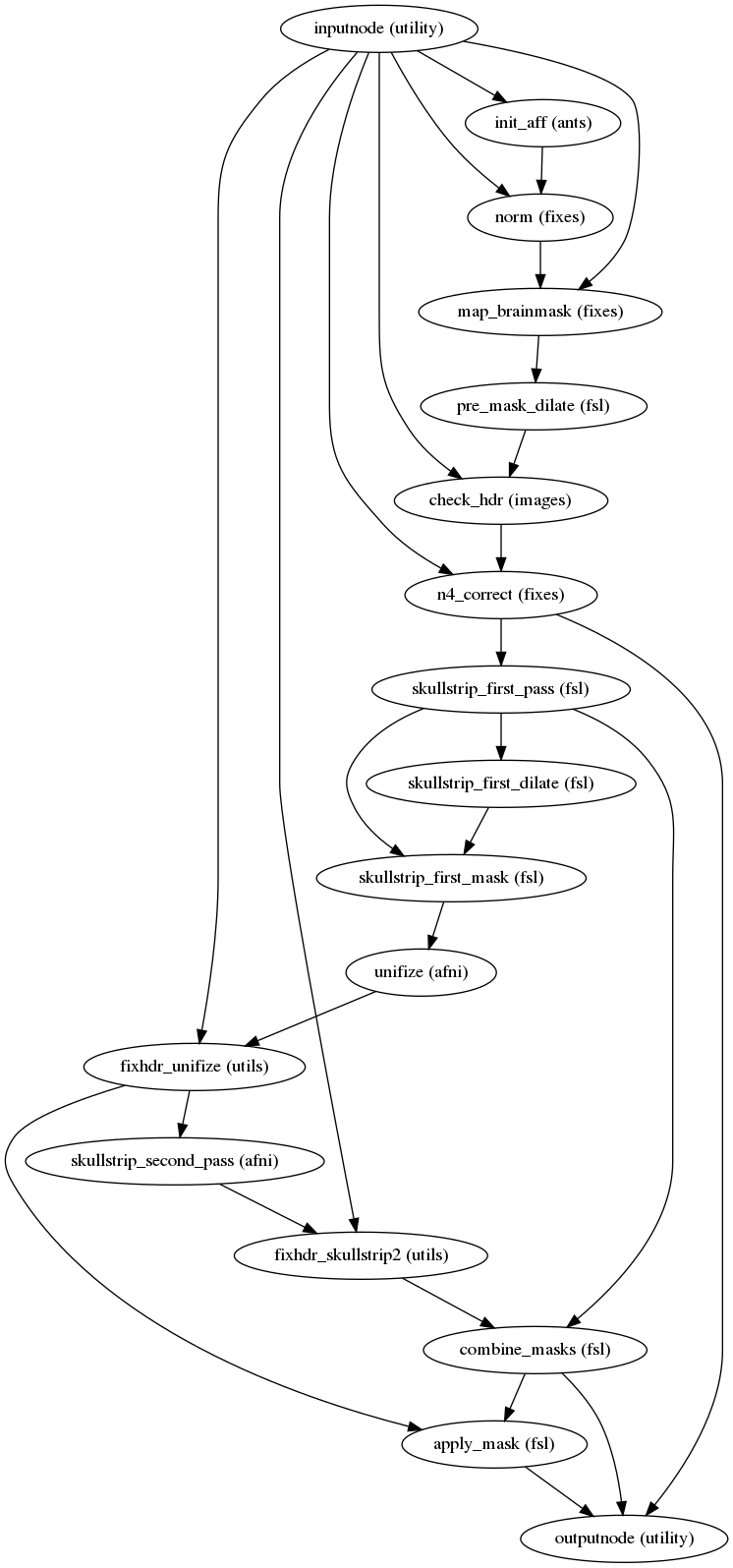
(Source code, png, svg, pdf)
- Parameters
name (str) – Name of workflow (default:
enhance_and_skullstrip_bold_wf)pre_mask (bool) – Indicates whether the
pre_maskinput will be set (and thus, step 1 should be skipped).omp_nthreads (int) – number of threads available to parallel nodes
- Inputs
in_file (str) – BOLD image (single volume)
pre_mask (bool) – A tentative brain mask to initialize the workflow (requires
pre_maskparameter setTrue).
- Outputs
bias_corrected_file (str) – the
in_fileafter N4BiasFieldCorrectionskull_stripped_file (str) – the
bias_corrected_fileafter skull-strippingmask_file (str) – mask of the skull-stripped input file
out_report (str) – reportlet for the skull-stripping
-
fmriprep.workflows.bold.util.init_skullstrip_bold_wf(name='skullstrip_bold_wf')[source]¶ Apply skull-stripping to a BOLD image.
It is intended to be used on an image that has previously been bias-corrected with
init_enhance_and_skullstrip_bold_wf()- Workflow Graph
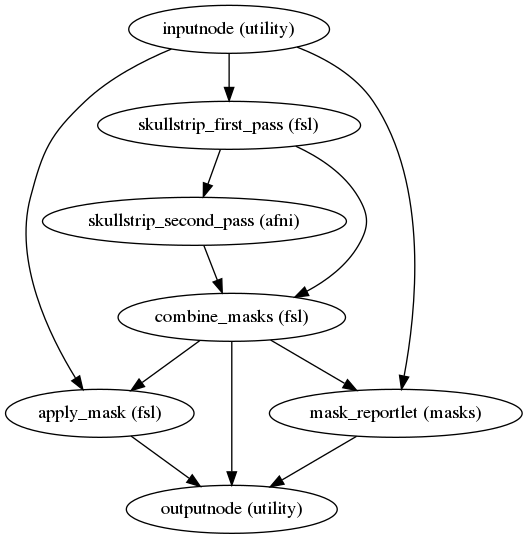
(Source code, png, svg, pdf)
- Inputs
in_file (str) – BOLD image (single volume)
- Outputs
skull_stripped_file (str) – the
in_fileafter skull-strippingmask_file (str) – mask of the skull-stripped input file
out_report (str) – reportlet for the skull-stripping
Head-Motion Estimation and Correction (HMC) of BOLD images¶
-
fmriprep.workflows.bold.hmc.init_bold_hmc_wf(mem_gb, omp_nthreads, name='bold_hmc_wf')[source]¶ Build a workflow to estimate head-motion parameters.
This workflow estimates the motion parameters to perform HMC over the input BOLD image.
- Workflow Graph
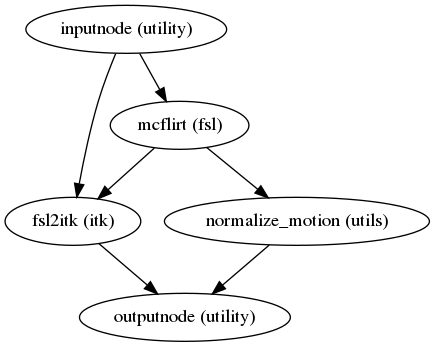
(Source code, png, svg, pdf)
- Parameters
mem_gb (float) – Size of BOLD file in GB
omp_nthreads (int) – Maximum number of threads an individual process may use
name (str) – Name of workflow (default:
bold_hmc_wf)
- Inputs
bold_file – BOLD series NIfTI file
raw_ref_image – Reference image to which BOLD series is motion corrected
- Outputs
xforms – ITKTransform file aligning each volume to
ref_imagemovpar_file – MCFLIRT motion parameters, normalized to SPM format (X, Y, Z, Rx, Ry, Rz)
Slice-Timing Correction (STC) of BOLD images¶
-
fmriprep.workflows.bold.stc.init_bold_stc_wf(metadata, name='bold_stc_wf')[source]¶ Create a workflow for STC.
This workflow performs STC over the input BOLD image.
- Workflow Graph
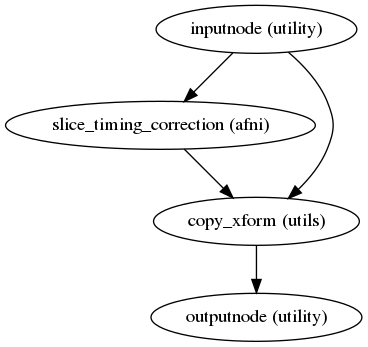
(Source code, png, svg, pdf)
- Parameters
metadata (dict) – BIDS metadata for BOLD file
name (str) – Name of workflow (default:
bold_stc_wf)
- Inputs
bold_file – BOLD series NIfTI file
skip_vols – Number of non-steady-state volumes detected at beginning of
bold_file
- Outputs
stc_file – Slice-timing corrected BOLD series NIfTI file
Generate T2* map from multi-echo BOLD images¶
-
fmriprep.workflows.bold.t2s.init_bold_t2s_wf(echo_times, mem_gb, omp_nthreads, t2s_coreg=False, name='bold_t2s_wf')[source]¶ Combine multiple echos of ME-EPI.
This workflow wraps the tedana T2* workflow to optimally combine multiple echos and derive a T2* map for optional use as a coregistration target. The following steps are performed:
HMC on individual echo files.
Compute the T2* map
Create an optimally combined ME-EPI time series
- Parameters
echo_times – list of TEs associated with each echo
mem_gb (float) – Size of BOLD file in GB
omp_nthreads (int) – Maximum number of threads an individual process may use
t2s_coreg (bool) – Use the calculated T2*-map for T2*-driven coregistration
name (str) – Name of workflow (default:
bold_t2s_wf)
- Inputs
bold_file – list of individual echo files
- Outputs
bold – the optimally combined time series for all supplied echos
bold_mask – the binarized, skull-stripped adaptive T2* map
bold_ref_brain – the adaptive T2* map
Registration workflows¶
-
fmriprep.workflows.bold.registration.init_bold_reg_wf(freesurfer, use_bbr, bold2t1w_dof, mem_gb, omp_nthreads, use_compression=True, write_report=True, name='bold_reg_wf')[source]¶ Build a workflow to run same-subject, BOLD-to-T1w image-registration.
Calculates the registration between a reference BOLD image and T1w-space using a boundary-based registration (BBR) cost function. If FreeSurfer-based preprocessing is enabled, the
bbregisterutility is used to align the BOLD images to the reconstructed subject, and the resulting transform is adjusted to target the T1 space. If FreeSurfer-based preprocessing is disabled, FSL FLIRT is used with the BBR cost function to directly target the T1 space.- Workflow Graph
(Source code, png, svg, pdf)
- Parameters
freesurfer (bool) – Enable FreeSurfer functional registration (bbregister)
use_bbr (bool or None) – Enable/disable boundary-based registration refinement. If
None, test BBR result for distortion before accepting.bold2t1w_dof (6, 9 or 12) – Degrees-of-freedom for BOLD-T1w registration
mem_gb (float) – Size of BOLD file in GB
omp_nthreads (int) – Maximum number of threads an individual process may use
name (str) – Name of workflow (default:
bold_reg_wf)use_compression (bool) – Save registered BOLD series as
.nii.gzuse_fieldwarp (bool) – Include SDC warp in single-shot transform from BOLD to T1
write_report (bool) – Whether a reportlet should be stored
- Inputs
ref_bold_brain – Reference image to which BOLD series is aligned If
fieldwarp == True,ref_bold_brainshould be unwarpedt1w_brain – Skull-stripped
t1w_preproct1w_dseg – Segmentation of preprocessed structural image, including gray-matter (GM), white-matter (WM) and cerebrospinal fluid (CSF)
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
fsnative2t1w_xfm – LTA-style affine matrix translating from FreeSurfer-conformed subject space to T1w
- Outputs
itk_bold_to_t1 – Affine transform from
ref_bold_brainto T1 space (ITK format)itk_t1_to_bold – Affine transform from T1 space to BOLD space (ITK format)
fallback – Boolean indicating whether BBR was rejected (mri_coreg registration returned)
-
fmriprep.workflows.bold.registration.init_bold_t1_trans_wf(freesurfer, mem_gb, omp_nthreads, multiecho=False, use_fieldwarp=False, use_compression=True, name='bold_t1_trans_wf')[source]¶ Co-register the reference BOLD image to T1w-space.
The workflow uses BBR.
- Workflow Graph
(Source code, png, svg, pdf)
- Parameters
freesurfer (bool) – Enable FreeSurfer functional registration (bbregister)
use_fieldwarp (bool) – Include SDC warp in single-shot transform from BOLD to T1
multiecho (bool) – If multiecho data was supplied, HMC already performed
mem_gb (float) – Size of BOLD file in GB
omp_nthreads (int) – Maximum number of threads an individual process may use
use_compression (bool) – Save registered BOLD series as
.nii.gzname (str) – Name of workflow (default:
bold_reg_wf)
- Inputs
name_source – BOLD series NIfTI file Used to recover original information lost during processing
ref_bold_brain – Reference image to which BOLD series is aligned If
fieldwarp == True,ref_bold_brainshould be unwarpedref_bold_mask – Skull-stripping mask of reference image
t1w_brain – Skull-stripped bias-corrected structural template image
t1w_mask – Mask of the skull-stripped template image
t1w_aseg – FreeSurfer’s
aseg.mgzatlas projected into the T1w reference (only ifrecon-allwas run).t1w_aparc – FreeSurfer’s
aparc+aseg.mgzatlas projected into the T1w reference (only ifrecon-allwas run).bold_split – Individual 3D BOLD volumes, not motion corrected
hmc_xforms – List of affine transforms aligning each volume to
ref_imagein ITK formatitk_bold_to_t1 – Affine transform from
ref_bold_brainto T1 space (ITK format)fieldwarp – a DFM in ITK format
- Outputs
bold_t1 – Motion-corrected BOLD series in T1 space
bold_t1_ref – Reference, contrast-enhanced summary of the motion-corrected BOLD series in T1w space
bold_mask_t1 – BOLD mask in T1 space
bold_aseg_t1 – FreeSurfer’s
aseg.mgzatlas, in T1w-space at the BOLD resolution (only ifrecon-allwas run).bold_aparc_t1 – FreeSurfer’s
aparc+aseg.mgzatlas, in T1w-space at the BOLD resolution (only ifrecon-allwas run).
-
fmriprep.workflows.bold.registration.init_bbreg_wf(use_bbr, bold2t1w_dof, omp_nthreads, name='bbreg_wf')[source]¶ Build a workflow to run FreeSurfer’s
bbregister.This workflow uses FreeSurfer’s
bbregisterto register a BOLD image to a T1-weighted structural image.It is a counterpart to
init_fsl_bbr_wf(), which performs the same task using FSL’s FLIRT with a BBR cost function. Theuse_bbroption permits a high degree of control over registration. IfFalse, standard, affine coregistration will be performed using FreeSurfer’smri_coregtool. IfTrue,bbregisterwill be seeded with the initial transform found bymri_coreg(equivalent to runningbbregister --init-coreg). IfNone, afterbbregisteris run, the resulting affine transform will be compared to the initial transform found bymri_coreg. Excessive deviation will result in rejecting the BBR refinement and accepting the original, affine registration.- Workflow Graph
(Source code, png, svg, pdf)
- Parameters
use_bbr (bool or None) – Enable/disable boundary-based registration refinement. If
None, test BBR result for distortion before accepting.bold2t1w_dof (6, 9 or 12) – Degrees-of-freedom for BOLD-T1w registration
name (str, optional) – Workflow name (default: bbreg_wf)
- Inputs
in_file – Reference BOLD image to be registered
fsnative2t1w_xfm – FSL-style affine matrix translating from FreeSurfer T1.mgz to T1w
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID (must have folder in SUBJECTS_DIR)
t1w_brain – Unused (see
init_fsl_bbr_wf())t1w_dseg – Unused (see
init_fsl_bbr_wf())
- Outputs
itk_bold_to_t1 – Affine transform from
ref_bold_brainto T1 space (ITK format)itk_t1_to_bold – Affine transform from T1 space to BOLD space (ITK format)
out_report – Reportlet for assessing registration quality
fallback – Boolean indicating whether BBR was rejected (mri_coreg registration returned)
-
fmriprep.workflows.bold.registration.init_fsl_bbr_wf(use_bbr, bold2t1w_dof, name='fsl_bbr_wf')[source]¶ Build a workflow to run FSL’s
flirt.This workflow uses FSL FLIRT to register a BOLD image to a T1-weighted structural image, using a boundary-based registration (BBR) cost function. It is a counterpart to
init_bbreg_wf(), which performs the same task using FreeSurfer’sbbregister.The
use_bbroption permits a high degree of control over registration. IfFalse, standard, rigid coregistration will be performed by FLIRT. IfTrue, FLIRT-BBR will be seeded with the initial transform found by the rigid coregistration. IfNone, after FLIRT-BBR is run, the resulting affine transform will be compared to the initial transform found by FLIRT. Excessive deviation will result in rejecting the BBR refinement and accepting the original, affine registration.- Workflow Graph
(Source code, png, svg, pdf)
- Parameters
use_bbr (bool or None) – Enable/disable boundary-based registration refinement. If
None, test BBR result for distortion before accepting.bold2t1w_dof (6, 9 or 12) – Degrees-of-freedom for BOLD-T1w registration
name (str, optional) – Workflow name (default: fsl_bbr_wf)
- Inputs
in_file – Reference BOLD image to be registered
t1w_brain – Skull-stripped T1-weighted structural image
t1w_dseg – FAST segmentation of
t1w_brainfsnative2t1w_xfm – Unused (see
init_bbreg_wf())subjects_dir – Unused (see
init_bbreg_wf())subject_id – Unused (see
init_bbreg_wf())
- Outputs
itk_bold_to_t1 – Affine transform from
ref_bold_brainto T1w space (ITK format)itk_t1_to_bold – Affine transform from T1 space to BOLD space (ITK format)
out_report – Reportlet for assessing registration quality
fallback – Boolean indicating whether BBR was rejected (rigid FLIRT registration returned)
Resampling workflows¶
-
fmriprep.workflows.bold.resampling.init_bold_surf_wf(mem_gb, output_spaces, medial_surface_nan, name='bold_surf_wf')[source]¶ Sample functional images to FreeSurfer surfaces.
For each vertex, the cortical ribbon is sampled at six points (spaced 20% of thickness apart) and averaged. Outputs are in GIFTI format.
- Workflow Graph
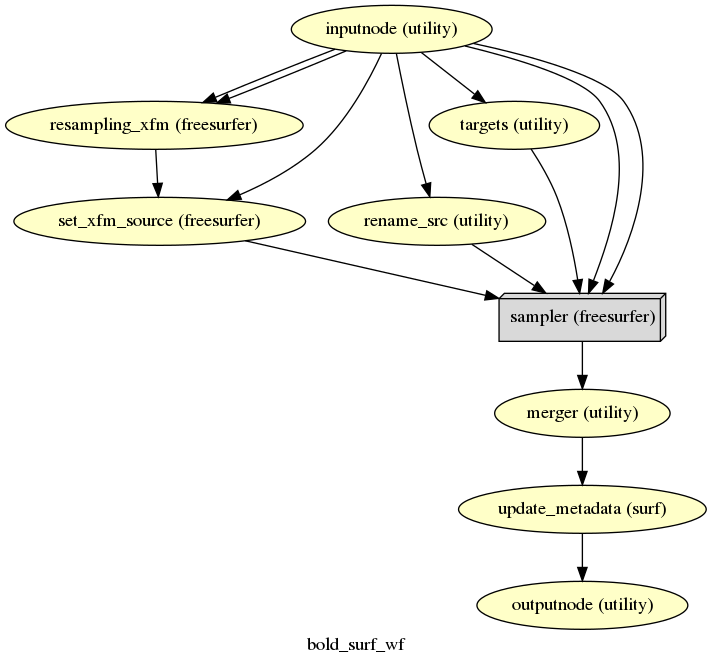
(Source code, png, svg, pdf)
- Parameters
output_spaces (list) – List of output spaces functional images are to be resampled to Target spaces beginning with
fswill be selected for resampling, such asfsaverageor related template spaces If the list containsfsnative, images will be resampled to the individual subject’s native surfacemedial_surface_nan (bool) – Replace medial wall values with NaNs on functional GIFTI files
- Inputs
source_file – Motion-corrected BOLD series in T1 space
t1w_preproc – Bias-corrected structural template image
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
t1w2fsnative_xfm – LTA-style affine matrix translating from T1w to FreeSurfer-conformed subject space
- Outputs
surfaces – BOLD series, resampled to FreeSurfer surfaces
-
fmriprep.workflows.bold.resampling.init_bold_std_trans_wf(freesurfer, mem_gb, omp_nthreads, standard_spaces, name='bold_std_trans_wf', use_compression=True, use_fieldwarp=False)[source]¶ Sample fMRI into standard space with a single-step resampling of the original BOLD series.
Important
This workflow provides two outputnodes. One output node (with name
poutputnode) will be parameterized in a Nipype sense (see Nipype iterables), and a second node (outputnode) will collapse the parameterized outputs into synchronous lists of the output fields listed below.- Workflow Graph
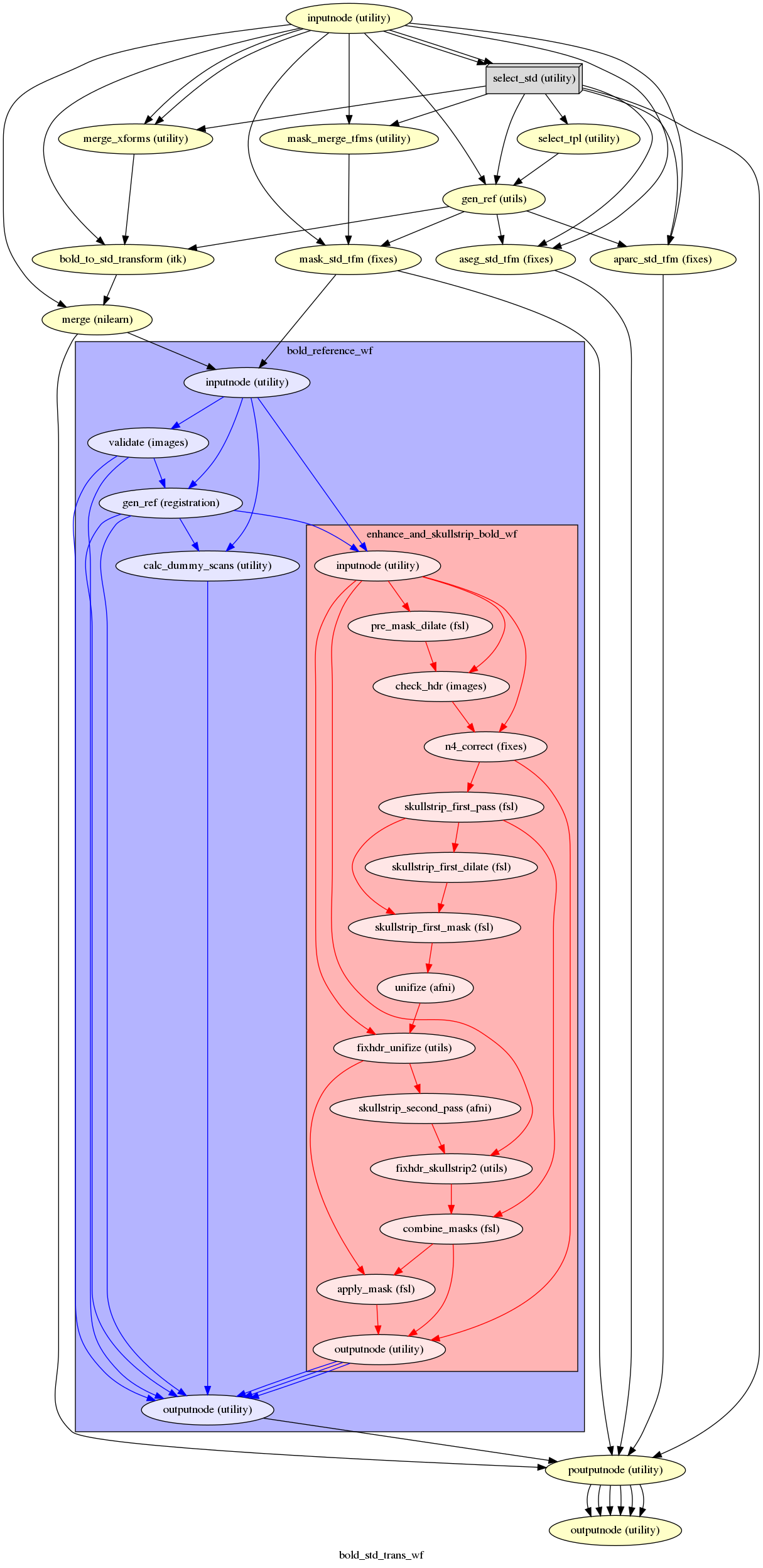
(Source code, png, svg, pdf)
- Parameters
freesurfer (bool) – Whether to generate FreeSurfer’s aseg/aparc segmentations on BOLD space.
mem_gb (float) – Size of BOLD file in GB
omp_nthreads (int) – Maximum number of threads an individual process may use
standard_spaces (OrderedDict) – Ordered dictionary where keys are TemplateFlow ID strings (e.g.,
MNI152Lin,MNI152NLin6Asym,MNI152NLin2009cAsym, orfsLR), or paths pointing to custom templates organized in a TemplateFlow-like structure. Values of the dictionary aggregate modifiers (e.g., the value for the keyMNI152Lincould be{'resolution': 2}if one wants the resampling to be done on the 2mm resolution version of the selected template).name (str) – Name of workflow (default:
bold_std_trans_wf)use_compression (bool) – Save registered BOLD series as
.nii.gzuse_fieldwarp (bool) – Include SDC warp in single-shot transform from BOLD to MNI
- Inputs
anat2std_xfm – List of anatomical-to-standard space transforms generated during spatial normalization.
bold_aparc – FreeSurfer’s
aparc+aseg.mgzatlas projected into the T1w reference (only ifrecon-allwas run).bold_aseg – FreeSurfer’s
aseg.mgzatlas projected into the T1w reference (only ifrecon-allwas run).bold_mask – Skull-stripping mask of reference image
bold_split – Individual 3D volumes, not motion corrected
fieldwarp – a DFM in ITK format
hmc_xforms – List of affine transforms aligning each volume to
ref_imagein ITK formatitk_bold_to_t1 – Affine transform from
ref_bold_brainto T1 space (ITK format)name_source – BOLD series NIfTI file Used to recover original information lost during processing
templates – List of templates that were applied as targets during spatial normalization.
- Outputs
bold_std – BOLD series, resampled to template space
bold_std_ref – Reference, contrast-enhanced summary of the BOLD series, resampled to template space
bold_mask_std – BOLD series mask in template space
bold_aseg_std – FreeSurfer’s
aseg.mgzatlas, in template space at the BOLD resolution (only ifrecon-allwas run)bold_aparc_std – FreeSurfer’s
aparc+aseg.mgzatlas, in template space at the BOLD resolution (only ifrecon-allwas run)templates – Template identifiers synchronized correspondingly to previously described outputs.
-
fmriprep.workflows.bold.resampling.init_bold_preproc_trans_wf(mem_gb, omp_nthreads, name='bold_preproc_trans_wf', use_compression=True, use_fieldwarp=False, split_file=False, interpolation='LanczosWindowedSinc')[source]¶ Resample in native (original) space.
This workflow resamples the input fMRI in its native (original) space in a “single shot” from the original BOLD series.
- Workflow Graph
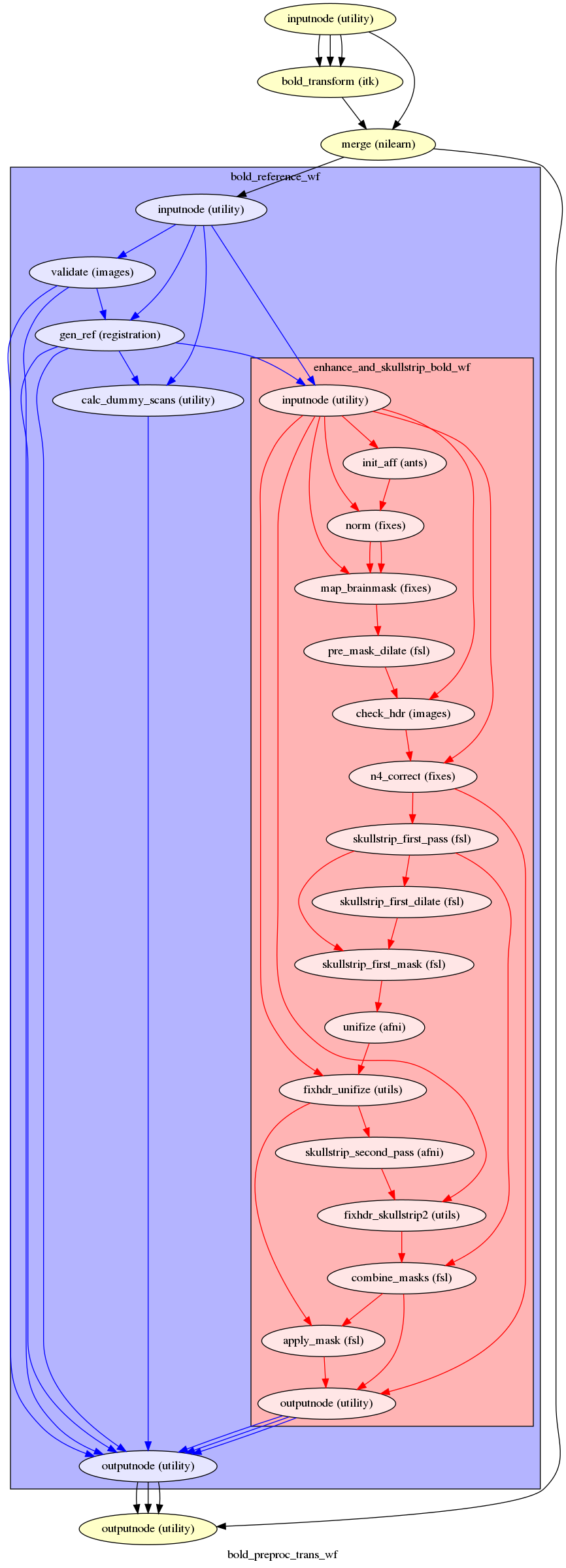
(Source code, png, svg, pdf)
- Parameters
mem_gb (float) – Size of BOLD file in GB
omp_nthreads (int) – Maximum number of threads an individual process may use
name (str) – Name of workflow (default:
bold_std_trans_wf)use_compression (bool) – Save registered BOLD series as
.nii.gzuse_fieldwarp (bool) – Include SDC warp in single-shot transform from BOLD to MNI
split_file (bool) – Whether the input file should be splitted (it is a 4D file) or it is a list of 3D files (default
False, do not split)interpolation (str) – Interpolation type to be used by ANTs’
applyTransforms(default'LanczosWindowedSinc')
- Inputs
bold_file – Individual 3D volumes, not motion corrected
bold_mask – Skull-stripping mask of reference image
name_source – BOLD series NIfTI file Used to recover original information lost during processing
hmc_xforms – List of affine transforms aligning each volume to
ref_imagein ITK formatfieldwarp – a DFM in ITK format
- Outputs
bold – BOLD series, resampled in native space, including all preprocessing
bold_mask – BOLD series mask calculated with the new time-series
bold_ref – BOLD reference image: an average-like 3D image of the time-series
bold_ref_brain – Same as
bold_ref, but once the brain mask has been applied
Calculate BOLD confounds¶
-
fmriprep.workflows.bold.confounds.init_bold_confs_wf(mem_gb, metadata, regressors_all_comps, regressors_dvars_th, regressors_fd_th, name='bold_confs_wf')[source]¶ Build a workflow to generate and write out confounding signals.
This workflow calculates confounds for a BOLD series, and aggregates them into a TSV file, for use as nuisance regressors in a GLM. The following confounds are calculated, with column headings in parentheses:
Region-wise average signal (
csf,white_matter,global_signal)DVARS - original and standardized variants (
dvars,std_dvars)Framewise displacement, based on head-motion parameters (
framewise_displacement)Temporal CompCor (
t_comp_cor_XX)Anatomical CompCor (
a_comp_cor_XX)Cosine basis set for high-pass filtering w/ 0.008 Hz cut-off (
cosine_XX)Non-steady-state volumes (
non_steady_state_XX)Estimated head-motion parameters, in mm and rad (
trans_x,trans_y,trans_z,rot_x,rot_y,rot_z)
Prior to estimating aCompCor and tCompCor, non-steady-state volumes are censored and high-pass filtered using a DCT basis. The cosine basis, as well as one regressor per censored volume, are included for convenience.
- Workflow Graph
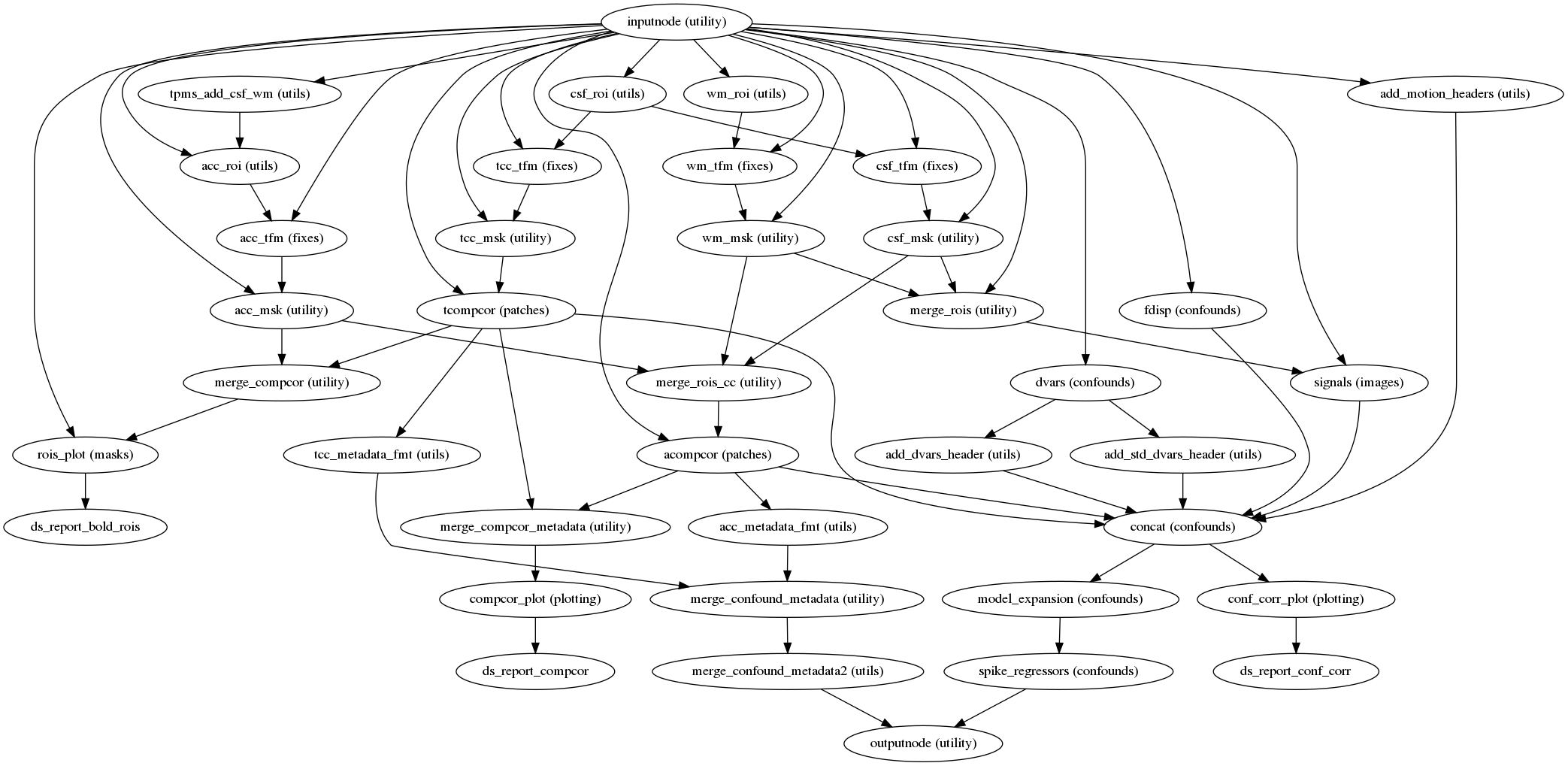
(Source code, png, svg, pdf)
- Parameters
mem_gb (float) – Size of BOLD file in GB - please note that this size should be calculated after resamplings that may extend the FoV
metadata (dict) – BIDS metadata for BOLD file
name (str) – Name of workflow (default:
bold_confs_wf)regressors_all_comps (bool) – Indicates whether CompCor decompositions should return all components instead of the minimal number of components necessary to explain 50 percent of the variance in the decomposition mask.
regressors_dvars_th – Criterion for flagging DVARS outliers
regressors_fd_th – Criterion for flagging framewise displacement outliers
- Inputs
bold – BOLD image, after the prescribed corrections (STC, HMC and SDC) when available.
bold_mask – BOLD series mask
movpar_file – SPM-formatted motion parameters file
skip_vols – number of non steady state volumes
t1w_mask – Mask of the skull-stripped template image
t1w_tpms – List of tissue probability maps in T1w space
t1_bold_xform – Affine matrix that maps the T1w space into alignment with the native BOLD space
- Outputs
confounds_file – TSV of all aggregated confounds
rois_report – Reportlet visualizing white-matter/CSF mask used for aCompCor, the ROI for tCompCor and the BOLD brain mask.
confounds_metadata – Confounds metadata dictionary.
-
fmriprep.workflows.bold.confounds.init_ica_aroma_wf(metadata, mem_gb, omp_nthreads, name='ica_aroma_wf', susan_fwhm=6.0, err_on_aroma_warn=False, aroma_melodic_dim=-200, use_fieldwarp=True)[source]¶ Build a workflow that runs ICA-AROMA.
This workflow wraps ICA-AROMA to identify and remove motion-related independent components from a BOLD time series.
The following steps are performed:
Remove non-steady state volumes from the bold series.
Smooth data using FSL susan, with a kernel width FWHM=6.0mm.
Run FSL melodic outside of ICA-AROMA to generate the report
Run ICA-AROMA
Aggregate identified motion components (aggressive) to TSV
Return
classified_motion_ICsandmelodic_mixfor user to complete non-aggressive denoising in T1w spaceCalculate ICA-AROMA-identified noise components (columns named
AROMAAggrCompXX)
Additionally, non-aggressive denoising is performed on the BOLD series resampled into MNI space.
There is a current discussion on whether other confounds should be extracted before or after denoising here.
- Workflow Graph
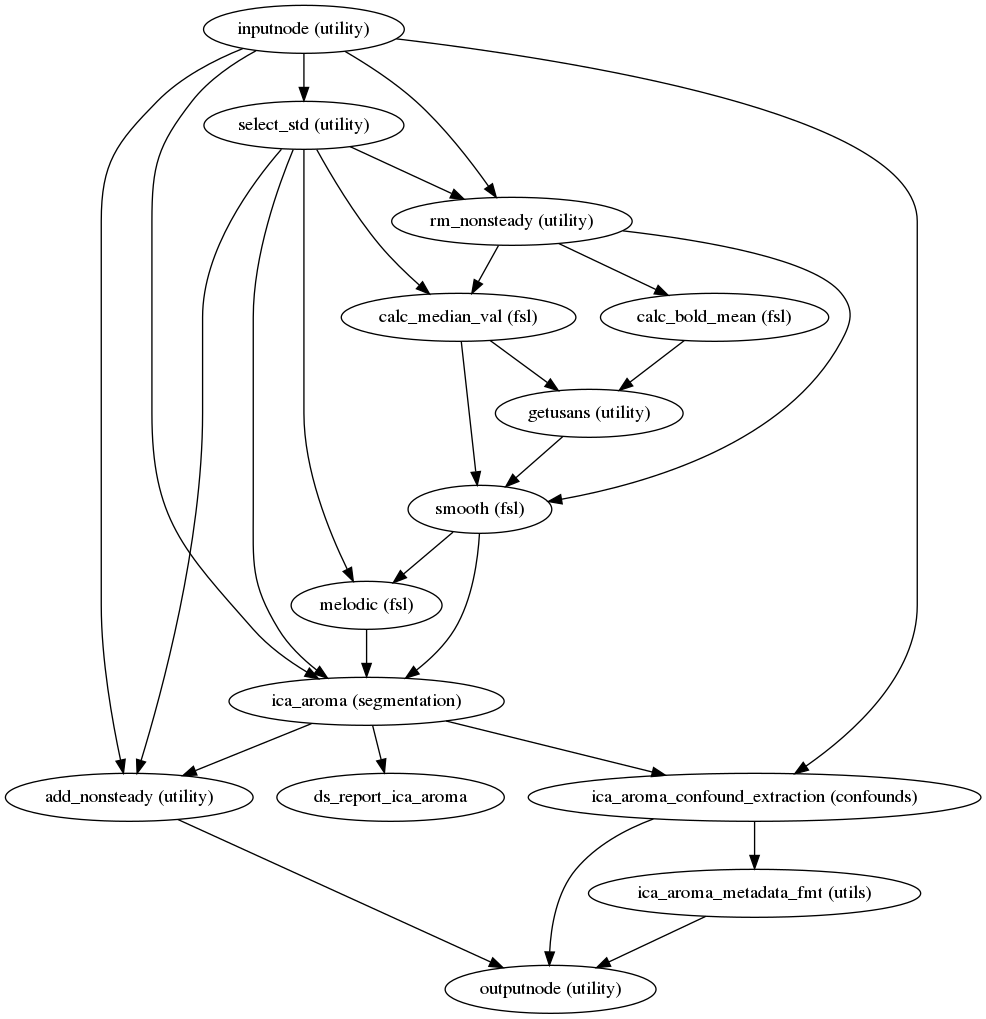
(Source code, png, svg, pdf)
- Parameters
standard_spaces (str) – Spatial normalization template used as target when that registration step was previously calculated with
init_bold_reg_wf(). The template must be one of the MNI templates (fMRIPrep usesMNI152NLin2009cAsymby default).metadata (dict) – BIDS metadata for BOLD file
mem_gb (float) – Size of BOLD file in GB
omp_nthreads (int) – Maximum number of threads an individual process may use
name (str) – Name of workflow (default:
bold_tpl_trans_wf)susan_fwhm (float) – Kernel width (FWHM in mm) for the smoothing step with FSL
susan(default: 6.0mm)use_fieldwarp (bool) – Include SDC warp in single-shot transform from BOLD to MNI
err_on_aroma_warn (bool) – Do not fail on ICA-AROMA errors
aroma_melodic_dim (int) – Set the dimensionality of the MELODIC ICA decomposition. Negative numbers set a maximum on automatic dimensionality estimation. Positive numbers set an exact number of components to extract. (default: -200, i.e., estimate <=200 components)
- Inputs
itk_bold_to_t1 – Affine transform from
ref_bold_brainto T1 space (ITK format)anat2std_xfm – ANTs-compatible affine-and-warp transform file
name_source – BOLD series NIfTI file Used to recover original information lost during processing
skip_vols – number of non steady state volumes
bold_split – Individual 3D BOLD volumes, not motion corrected
bold_mask – BOLD series mask in template space
hmc_xforms – List of affine transforms aligning each volume to
ref_imagein ITK formatfieldwarp – a DFM in ITK format
movpar_file – SPM-formatted motion parameters file
- Outputs
aroma_confounds – TSV of confounds identified as noise by ICA-AROMA
aroma_noise_ics – CSV of noise components identified by ICA-AROMA
melodic_mix – FSL MELODIC mixing matrix
nonaggr_denoised_file – BOLD series with non-aggressive ICA-AROMA denoising applied
Fieldmap estimation and unwarping workflows¶
Automatic selection of the appropriate SDC method¶
If the dataset metadata indicate tha more than one field map acquisition is
IntendedFor (see BIDS Specification section 8.9) the following priority will
be used:
Table of behavior (fieldmap use-cases):
Fieldmaps found |
|
|
Action |
|---|---|---|---|
True |
True |
Fieldmaps + SyN |
|
True |
False |
Fieldmaps |
|
False |
True |
SyN |
|
False |
True |
False |
SyN |
False |
False |
False |
HMC only |
-
fmriprep.workflows.fieldmap.base.init_sdc_wf(fmaps, bold_meta, omp_nthreads=1, debug=False, fmap_bspline=False, fmap_demean=True)[source]¶ This workflow implements the heuristics to choose a SDC strategy. When no field map information is present within the BIDS inputs, the EXPERIMENTAL “fieldmap-less SyN” can be performed, using the
--use-synargument. When--force-synis specified, then the “fieldmap-less SyN” is always executed and reported despite of other fieldmaps available with higher priority. In the latter case (some sort of fieldmap(s) is available and--force-synis requested), then the SDC method applied is that with the highest priority.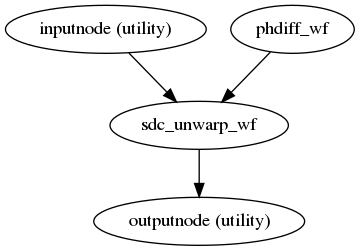
(Source code, png, svg, pdf)
Parameters
- fmapslist of pybids dicts
A list of dictionaries with the available fieldmaps (and their metadata using the key
'metadata'for the case of epi fieldmaps)- bold_metadict
BIDS metadata dictionary corresponding to the BOLD run
- omp_nthreadsint
Maximum number of threads an individual process may use
- fmap_bsplinebool
Experimental: Fit B-Spline field using least-squares
- fmap_demeanbool
Demean voxel-shift map during unwarp
- debugbool
Enable debugging outputs
- Inputs
- bold_ref
A BOLD reference calculated at a previous stage
- bold_ref_brain
Same as above, but brain-masked
- bold_mask
Brain mask for the BOLD run
- t1_brain
T1w image, brain-masked, for the fieldmap-less SyN method
- std2anat_xfm
List of standard-to-T1w transforms generated during spatial normalization (only for the fieldmap-less SyN method).
- templatestr
Name of template from which prior knowledge will be mapped into the subject’s T1w reference (only for the fieldmap-less SyN method)
- templatesstr
Name of templates that index the
std2anat_xfminput list (only for the fieldmap-less SyN method).
- Outputs
- bold_ref
An unwarped BOLD reference
- bold_mask
The corresponding new mask after unwarping
- bold_ref_brain
Brain-extracted, unwarped BOLD reference
- out_warp
The deformation field to unwarp the susceptibility distortions
- syn_bold_ref
If
--force-syn, an unwarped BOLD reference with this method (for reporting purposes)
Direct B0 mapping sequences¶
When the fieldmap is directly measured with a prescribed sequence (such as SE), we only need to calculate the corresponding B-Spline coefficients to adapt the fieldmap to the TOPUP tool. This procedure is described with more detail here.
This corresponds to the section 8.9.3 –fieldmap image (and one magnitude image)– of the BIDS specification.
-
fmriprep.workflows.fieldmap.fmap.init_fmap_wf(omp_nthreads, fmap_bspline, name='fmap_wf')[source]¶ Fieldmap workflow - when we have a sequence that directly measures the fieldmap we just need to mask it (using the corresponding magnitude image) to remove the noise in the surrounding air region, and ensure that units are Hz.
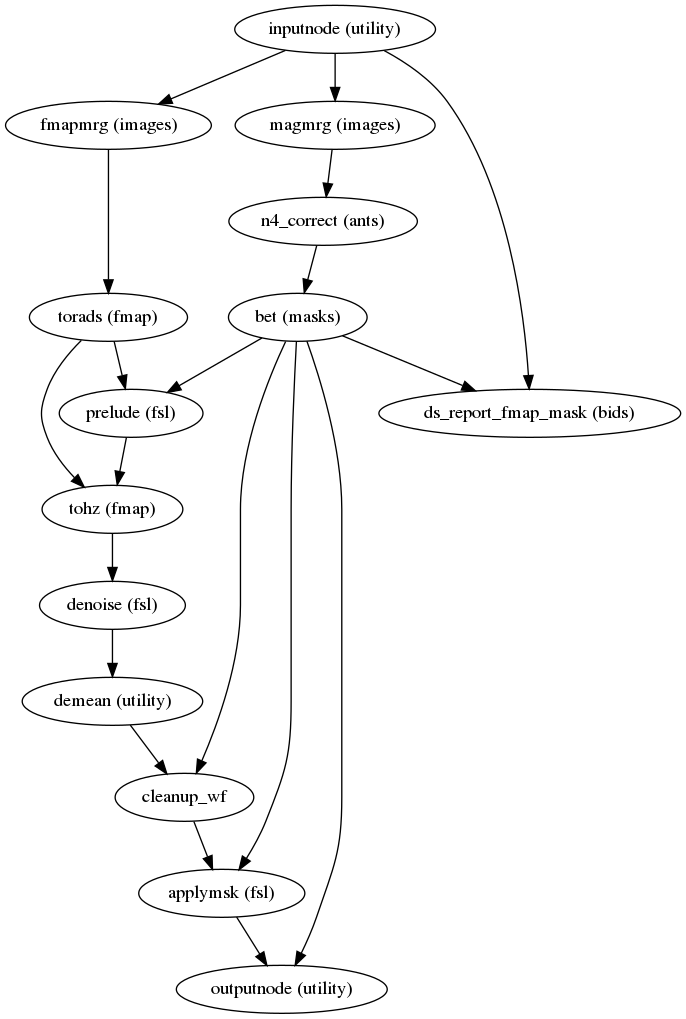
(Source code, png, svg, pdf)
Phase-difference B0 estimation¶
The field inhomogeneity inside the scanner (fieldmap) is proportional to the phase drift between two subsequent GRE sequence.
Fieldmap preprocessing workflow for fieldmap data structure 8.9.1 in BIDS 1.0.0: one phase diff and at least one magnitude image
-
fmriprep.workflows.fieldmap.phdiff.init_phdiff_wf(omp_nthreads, name='phdiff_wf')[source]¶ Estimates the fieldmap using a phase-difference image and one or more magnitude images corresponding to two or more GRE acquisitions. The original code was taken from nipype.
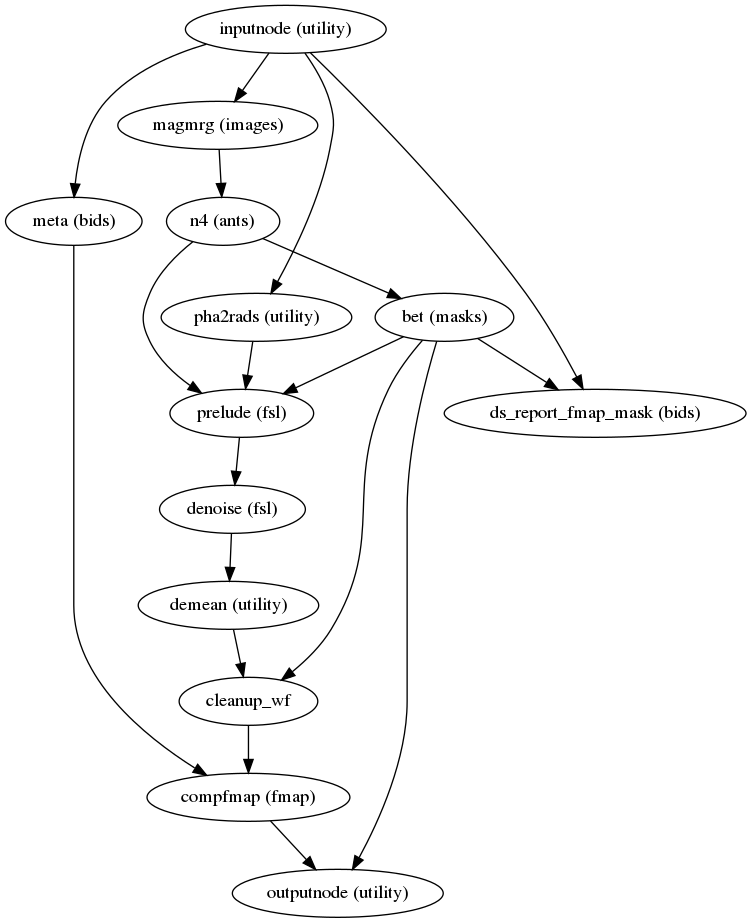
(Source code, png, svg, pdf)
Outputs:
outputnode.fmap_ref - The average magnitude image, skull-stripped outputnode.fmap_mask - The brain mask applied to the fieldmap outputnode.fmap - The estimated fieldmap in Hz
Phase Encoding POLARity (PEPOLAR) techniques¶
-
fmriprep.workflows.fieldmap.pepolar.init_pepolar_unwarp_wf(bold_meta, epi_fmaps, omp_nthreads=1, name='pepolar_unwarp_wf')[source]¶ This workflow takes in a set of EPI files with opposite phase encoding direction than the target file and calculates a displacements field (in other words, an ANTs-compatible warp file).
This procedure works if there is only one ‘_epi’ file is present (as long as it has the opposite phase encoding direction to the target file). The target file will be used to estimate the field distortion. However, if there is another ‘_epi’ file present with a matching phase encoding direction to the target it will be used instead.
Currently, different phase encoding dimension in the target file and the ‘_epi’ file(s) (for example ‘i’ and ‘j’) is not supported.
The warp field correcting for the distortions is estimated using AFNI’s 3dQwarp, with displacement estimation limited to the target file phase encoding direction.
It also calculates a new mask for the input dataset that takes into account the distortions.
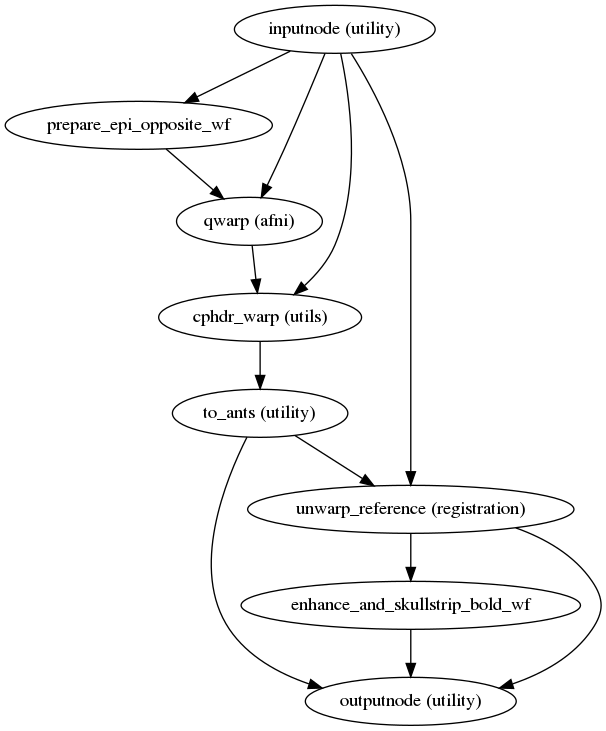
(Source code, png, svg, pdf)
Inputs
- in_reference
the reference image
- in_reference_brain
the reference image skullstripped
- in_mask
a brain mask corresponding to
in_reference
Outputs
- out_reference
the
in_referenceafter unwarping- out_reference_brain
the
in_referenceafter unwarping and skullstripping- out_warp
the corresponding DFM compatible with ANTs
- out_mask
mask of the unwarped input file
-
fmriprep.workflows.fieldmap.pepolar.init_prepare_epi_wf(omp_nthreads, name='prepare_epi_wf')[source]¶ This workflow takes in a set of EPI files with with the same phase encoding direction and returns a single 3D volume ready to be used in field distortion estimation.
The procedure involves: estimating a robust template using FreeSurfer’s ‘mri_robust_template’, bias field correction using ANTs N4BiasFieldCorrection and AFNI 3dUnifize, skullstripping using FSL BET and AFNI 3dAutomask, and rigid coregistration to the reference using ANTs.
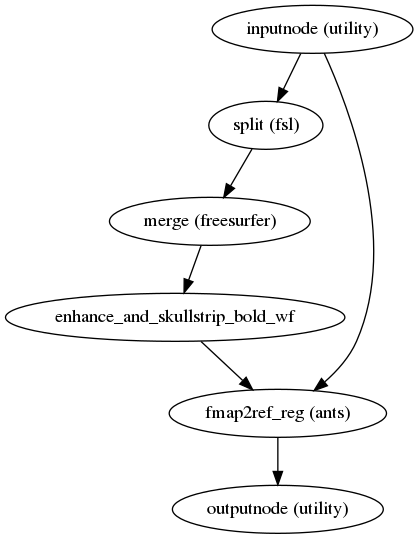
(Source code, png, svg, pdf)
Inputs
- fmaps
list of 3D or 4D NIfTI images
- ref_brain
coregistration reference (skullstripped and bias field corrected)
Outputs
- out_file
single 3D NIfTI file
Fieldmap-less estimation (experimental)¶
In the absence of direct measurements of fieldmap data, we provide an (experimental)
option to estimate the susceptibility distortion based on the ANTs symmetric
normalization (SyN) technique.
This feature may be enabled, using the --use-syn-sdc flag, and will only be
applied if fieldmaps are unavailable.
During the evaluation phase, the --force-syn flag will cause this estimation to
be performed in addition to fieldmap-based estimation, to permit the direct
comparison of the results of each technique.
Note that, even if --force-syn is given, the functional outputs of FMRIPREP will
be corrected using the fieldmap-based estimates.
Feedback will be enthusiastically received.
-
fmriprep.workflows.fieldmap.syn.init_syn_sdc_wf(omp_nthreads, bold_pe=None, atlas_threshold=3, name='syn_sdc_wf')[source]¶ This workflow takes a skull-stripped T1w image and reference BOLD image and estimates a susceptibility distortion correction warp, using ANTs symmetric normalization (SyN) and the average fieldmap atlas described in [Treiber2016].
SyN deformation is restricted to the phase-encoding (PE) direction. If no PE direction is specified, anterior-posterior PE is assumed.
SyN deformation is also restricted to regions that are expected to have a >3mm (approximately 1 voxel) warp, based on the fieldmap atlas.
This technique is a variation on those developed in [Huntenburg2014] and [Wang2017].
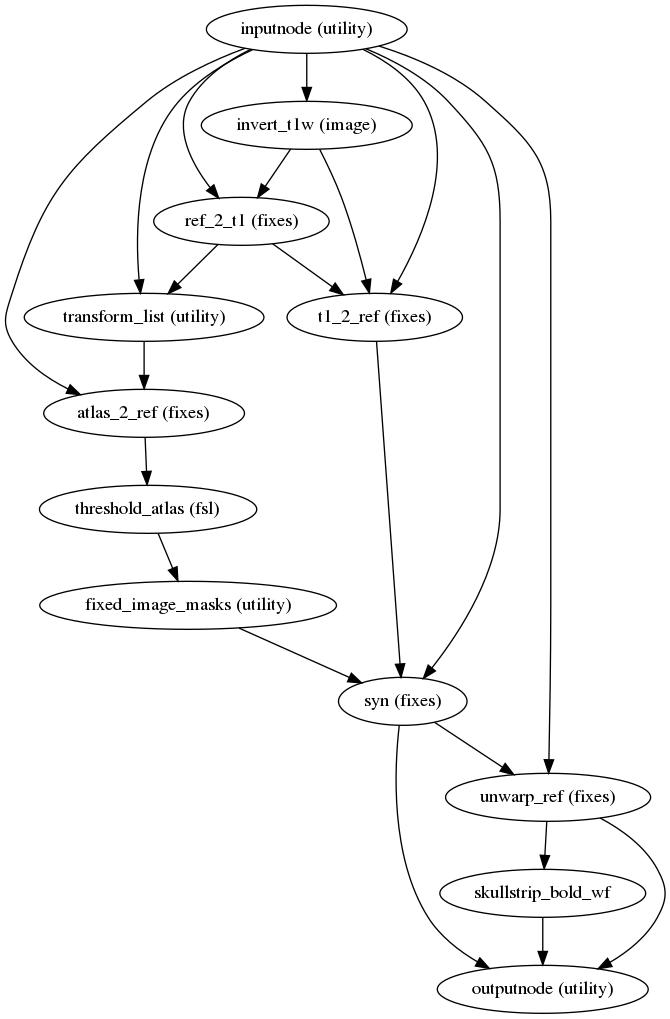
(Source code, png, svg, pdf)
Inputs
- bold_ref
reference image
- bold_ref_brain
skull-stripped reference image
- templatestr
Name of template targeted by
templateoutput space- t1_brain
skull-stripped, bias-corrected structural image
- std2anat_xfm
inverse registration transform of T1w image to MNI template
Outputs
- out_reference
the
bold_refimage after unwarping- out_reference_brain
the
bold_ref_brainimage after unwarping- out_warp
the corresponding DFM compatible with ANTs
- out_mask
mask of the unwarped input file
Unwarping¶
Abbreviations
- fmap
fieldmap
- VSM
voxel-shift map – a 3D nifti where displacements are in pixels (not mm)
- DFM
displacements field map – a nifti warp file compatible with ANTs (mm)
-
fmriprep.workflows.fieldmap.unwarp.init_sdc_unwarp_wf(omp_nthreads, fmap_demean, debug, name='sdc_unwarp_wf')[source]¶ Apply the warping given by a displacements fieldmap.
This workflow takes in a displacements fieldmap and calculates the corresponding displacements field (in other words, an ANTs-compatible warp file).
It also calculates a new mask for the input dataset that takes into account the distortions. The mask is restricted to the field of view of the fieldmap since outside of it corrections could not be performed.
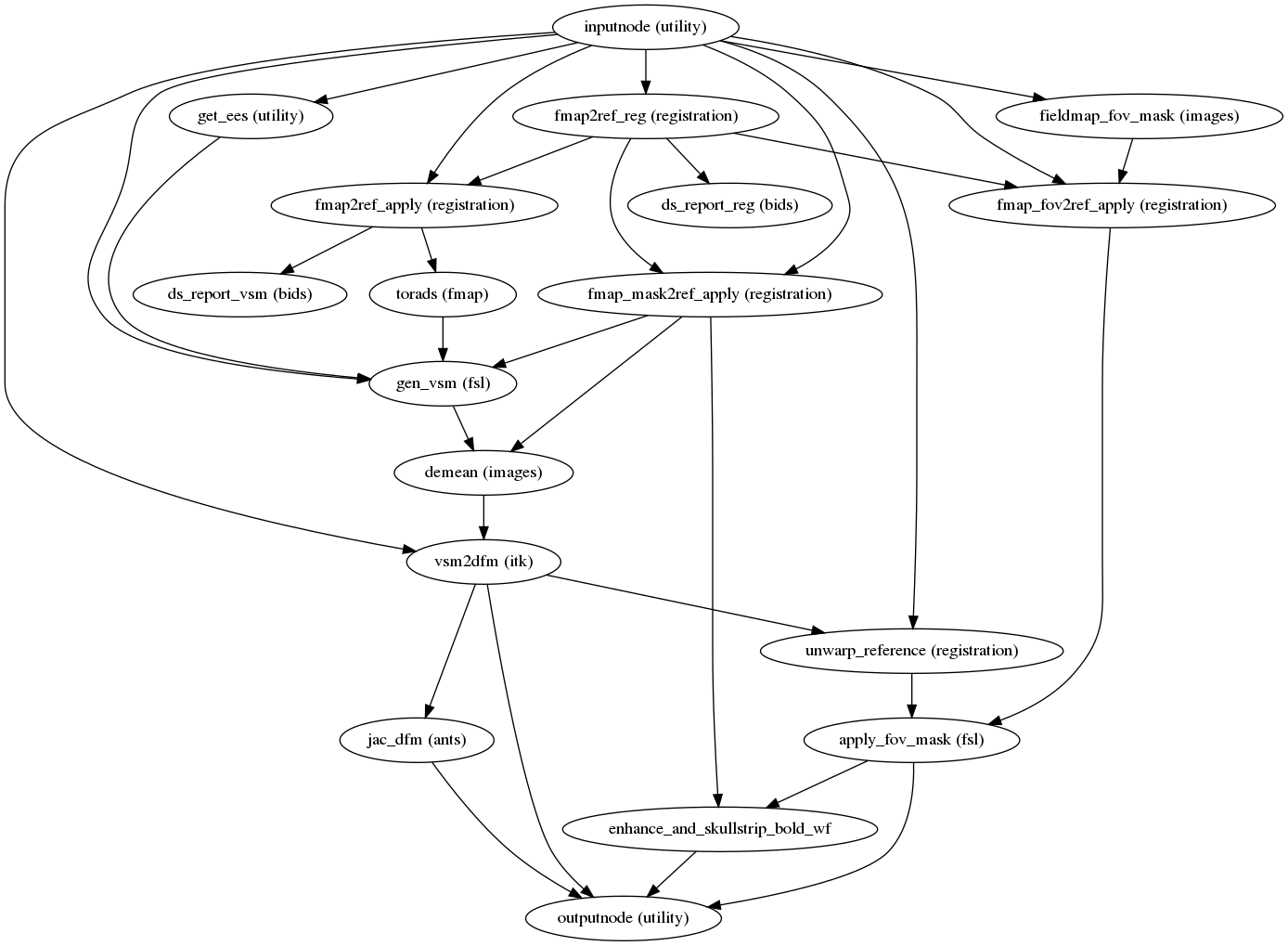
(Source code, png, svg, pdf)
Inputs
- in_reference
the reference image
- in_reference_brain
the reference image (skull-stripped)
- in_mask
a brain mask corresponding to
in_reference- metadata
metadata associated to the
in_referenceEPI input- fmap
the fieldmap in Hz
- fmap_ref
the reference (anatomical) image corresponding to
fmap- fmap_mask
a brain mask corresponding to
fmap
Outputs
- out_reference
the
in_referenceafter unwarping- out_reference_brain
the
in_referenceafter unwarping and skullstripping- out_warp
the corresponding DFM compatible with ANTs
- out_jacobian
the jacobian of the field (for drop-out alleviation)
- out_mask
mask of the unwarped input file
-
fmriprep.workflows.fieldmap.unwarp.init_fmap_unwarp_report_wf(name='fmap_unwarp_report_wf', forcedsyn=False)[source]¶ Save a reportlet showing how SDC unwarping performed.
This workflow generates and saves a reportlet showing the effect of fieldmap unwarping a BOLD image.
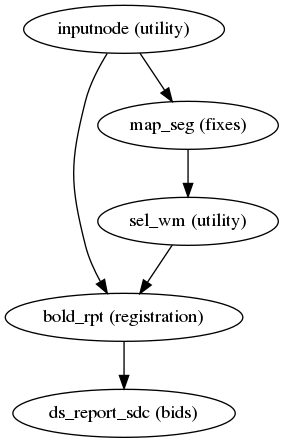
(Source code, png, svg, pdf)
Parameters
- namestr, optional
Workflow name (default: fmap_unwarp_report_wf)
- forcedsynbool, optional
Whether SyN-SDC was forced.
Inputs
- in_pre
Reference image, before unwarping
- in_post
Reference image, after unwarping
- in_seg
Segmentation of preprocessed structural image, including gray-matter (GM), white-matter (WM) and cerebrospinal fluid (CSF)
- in_xfm
Affine transform from T1 space to BOLD space (ITK format)