Developers - API
The NiPreps community and contributing guidelines
fMRIPrep is a NiPreps application, and abides by the NiPreps Community guidelines. Please, make sure you have read and understood all the documentation provided in the NiPreps portal before you get started.
Setting up your development environment
We believe that fMRIPrep must be free to use, inspect, and critique. Correspondingly, you should be free to modify our software to improve it or adapt it to new use cases and we especially welcome contributions to improve it or its documentation.
We actively direct efforts into making the scrutiny and improvement processes as easy as possible. As part of such efforts, we maintain some tips and guidelines for developers to help minimize your burden if you want to modify the software.
Internal configuration system
A Python module to maintain unique, run-wide fMRIPrep settings.
This module implements the memory structures to keep a consistent, singleton config.
Settings are passed across processes via filesystem, and a copy of the settings for
each run and subject is left under
<fmriprep_dir>/sub-<participant_id>/log/<run_unique_id>/fmriprep.toml.
Settings are stored using ToML.
The module has a to_filename() function to allow writing out
the settings to hard disk in ToML format, which looks like:
[environment]
cpu_count = 8
exec_env = "posix"
free_mem = 2.2
overcommit_policy = "heuristic"
overcommit_limit = "50%"
nipype_version = "1.5.0"
templateflow_version = "0.4.2"
version = "20.0.1"
[execution]
bids_dir = "ds000005/"
bids_description_hash = "5d42e27751bbc884eca87cb4e62b9a0cca0cd86f8e578747fe89b77e6c5b21e5"
boilerplate_only = false
fs_license_file = "/opt/freesurfer/license.txt"
fs_subjects_dir = "/opt/freesurfer/subjects"
log_dir = "/home/oesteban/tmp/fmriprep-ds005/out/fmriprep/logs"
log_level = 40
low_mem = false
md_only_boilerplate = false
notrack = true
output_dir = "/tmp"
output_spaces = "MNI152NLin2009cAsym:res-2 MNI152NLin2009cAsym:res-native fsaverage:den-10k fsaverage:den-30k"
reports_only = false
run_uuid = "20200306-105302_d365772b-fd60-4741-a722-372c2f558b50"
participant_label = [ "01",]
templateflow_home = "~/.cache/templateflow"
work_dir = "work/"
write_graph = false
[workflow]
anat_only = false
aroma_err_on_warn = false
aroma_melodic_dim = -200
bold2t1w_dof = 6
fmap_bspline = false
force_syn = false
hires = true
ignore = []
longitudinal = false
medial_surface_nan = false
project_goodvoxels = false
regressors_all_comps = false
regressors_dvars_th = 1.5
regressors_fd_th = 0.5
run_reconall = true
skull_strip_fixed_seed = false
skull_strip_template = "OASIS30ANTs"
t2s_coreg = false
use_aroma = false
[nipype]
crashfile_format = "txt"
get_linked_libs = false
memory_gb = 32
nprocs = 8
omp_nthreads = 8
plugin = "MultiProc"
resource_monitor = false
stop_on_first_crash = false
[nipype.plugin_args]
maxtasksperchild = 1
raise_insufficient = false
[execution.bids_filters.t1w]
reconstruction = "<Query.NONE: 1>"
[execution.bids_filters.t2w]
reconstruction = "<Query.NONE: 1>"
This config file is used to pass the settings across processes,
using the load() function.
Configuration sections
- class fmriprep.config.environment[source]
Read-only options regarding the platform and environment.
Crawls runtime descriptive settings (e.g., default FreeSurfer license, execution environment, nipype and fMRIPrep versions, etc.). The
environmentsection is not loaded in from file, only written out when settings are exported. This config section is useful when reporting issues, and these variables are tracked whenever the user does not opt-out using the--notrackargument.- cpu_count = 2
Number of available CPUs.
- exec_docker_version = None
Version of Docker Engine.
- exec_env = 'posix'
A string representing the execution platform.
- free_mem = 6.2
Free memory at start.
- nipype_version = '1.8.6'
Nipype’s current version.
- overcommit_limit = '50%'
Linux’s kernel virtual memory overcommit limits.
- overcommit_policy = 'heuristic'
Linux’s kernel virtual memory overcommit policy.
- templateflow_version = '24.0.0'
The TemplateFlow client version installed.
- version = '23.2.1'
fMRIPrep’s version.
- class fmriprep.config.execution[source]
Configure run-level settings.
- bids_database_dir = None[source]
Path to the directory containing SQLite database indices for the input BIDS dataset.
- bids_description_hash = None
Checksum (SHA256) of the
dataset_description.jsonof the BIDS dataset.
- bids_filters = None
A dictionary of BIDS selection filters.
- boilerplate_only = False
Only generate a boilerplate.
- country_code = 'CAN'
Country ISO code used by carbon trackers.
- debug = []
Debug mode(s).
- derivatives = []
Path(s) to search for pre-computed derivatives
- echo_idx = None
Select a particular echo for multi-echo EPI datasets.
- layout = None[source]
A
BIDSLayoutobject, seeinit().
- log_level = 25
Output verbosity.
- low_mem = None
Utilize uncompressed NIfTIs and other tricks to minimize memory allocation.
- md_only_boilerplate = False
Do not convert boilerplate from MarkDown to LaTex and HTML.
- me_output_echos = False
Output individual echo time series with slice, motion and susceptibility correction
- notrack = False
Do not collect telemetry information for fMRIPrep.
- output_layout = None
Layout of derivatives within output_dir.
- output_spaces = None
List of (non)standard spaces designated (with the
--output-spacesflag of the command line) as spatial references for outputs.
- participant_label = None
List of participant identifiers that are to be preprocessed.
- reports_only = False
Only build the reports, based on the reportlets found in a cached working directory.
- run_uuid = '20240306-164814_ca5524b4-e632-45dc-ae8f-f13be5ebdcb3'
Unique identifier of this particular run.
- sloppy = False
Run in sloppy mode (meaning, suboptimal parameters that minimize run-time).
- task_id = None
Select a particular task from all available in the dataset.
- templateflow_home = PosixPath('/home/docs/.cache/templateflow')[source]
The root folder of the TemplateFlow client.
- track_carbon = False
Tracks power draws using CodeCarbon package.
- work_dir = PosixPath('/home/docs/checkouts/readthedocs.org/user_builds/fmriprep/checkouts/stable/docs/work')[source]
Path to a working directory where intermediate results will be available.
- write_graph = False
Write out the computational graph corresponding to the planned preprocessing.
- class fmriprep.config.workflow[source]
Configure the particular execution graph of this workflow.
- anat_only = False
Execute the anatomical preprocessing only.
- aroma_err_on_warn = None
Cast AROMA warnings to errors.
- aroma_melodic_dim = None
Number of ICA components to be estimated by MELODIC (positive = exact, negative = maximum).
- bold2t1w_dof = None
Degrees of freedom of the BOLD-to-T1w registration steps.
- bold2t1w_init = 'register'
Whether to use standard coregistration (‘register’) or to initialize coregistration from the BOLD image-header (‘header’).
- cifti_output = None
Generate HCP Grayordinates, accepts either
'91k'(default) or'170k'.
- dummy_scans = None
Set a number of initial scans to be considered nonsteady states.
- fmap_bspline = None
Regularize fieldmaps with a field of B-Spline basis.
- fmap_demean = None
Remove the mean from fieldmaps.
- force_syn = None
Run fieldmap-less susceptibility-derived distortions estimation.
- hires = None
Run FreeSurfer
recon-allwith the-hiresflag.
- ignore = None
Ignore particular steps for fMRIPrep.
- level = None
Level of preprocessing to complete. One of [‘minimal’, ‘resampling’, ‘full’].
- longitudinal = False
Run FreeSurfer
recon-allwith the-logitudinalflag.
- me_t2s_fit_method = 'curvefit'
The method by which to estimate T2*/S0 for multi-echo data
- medial_surface_nan = None
Fill medial surface with NaNs when sampling.
- project_goodvoxels = False
Exclude voxels with locally high coefficient of variation from sampling.
- regressors_all_comps = None
Return all CompCor components.
- regressors_dvars_th = None
Threshold for DVARS.
- regressors_fd_th = None
Threshold for FD.
- run_msmsulc = True
Run Multimodal Surface Matching surface registration.
- run_reconall = True
Run FreeSurfer’s surface reconstruction.
- skull_strip_fixed_seed = False
Fix a seed for skull-stripping.
- skull_strip_t1w = 'force'
Skip brain extraction of the T1w image (default is
force, meaning that fMRIPrep will run brain extraction of the T1w).
- skull_strip_template = 'OASIS30ANTs'
Change default brain extraction template.
- slice_time_ref = 0.5
The time of the reference slice to correct BOLD values to, as a fraction acquisition time. 0 indicates the start, 0.5 the midpoint, and 1 the end of acquisition. The alias start corresponds to 0, and middle to 0.5. The default value is 0.5.
- spaces = None[source]
Keeps the
SpatialReferencesinstance keeping standard and nonstandard spaces.
- use_aroma = None
Run ICA-AROMA.
- use_bbr = None
Run boundary-based registration for BOLD-to-T1w registration.
- use_syn_sdc = None
Run fieldmap-less susceptibility-derived distortions estimation in the absence of any alternatives.
- class fmriprep.config.nipype[source]
Nipype settings.
- crashfile_format = 'txt'
The file format for crashfiles, either text (txt) or pickle (pklz).
- get_linked_libs = False
Run NiPype’s tool to enlist linked libraries for every interface.
- memory_gb = None
Estimation in GB of the RAM this workflow can allocate at any given time.
- nprocs = 2
Number of processes (compute tasks) that can be run in parallel (multiprocessing only).
- omp_nthreads = None
Number of CPUs a single process can access for multithreaded execution.
- plugin = 'MultiProc'
NiPype’s execution plugin.
- plugin_args = {'maxtasksperchild': 1, 'raise_insufficient': False}
Settings for NiPype’s execution plugin.
- remove_unnecessary_outputs = True
Clean up unused outputs after running
- resource_monitor = False
Enable resource monitor.
- stop_on_first_crash = True
Whether the workflow should stop or continue after the first error.
Usage
A config file is used to pass settings and collect information as the execution graph is built across processes.
from fmriprep import config
config_file = config.execution.work_dir / '.fmriprep.toml'
config.to_filename(config_file)
# Call build_workflow(config_file, retval) in a subprocess
with Manager() as mgr:
from .workflow import build_workflow
retval = mgr.dict()
p = Process(target=build_workflow, args=(str(config_file), retval))
p.start()
p.join()
config.load(config_file)
# Access configs from any code section as:
value = config.section.setting
Logging
Other responsibilities
The config is responsible for other conveniency actions.
Switching Python’s
multiprocessingto forkserver mode.Set up a filter for warnings as early as possible.
Automated I/O magic operations. Some conversions need to happen in the store/load processes (e.g., from/to
Path<->str,BIDSLayout, etc.)
- fmriprep.config.from_dict(settings, init=True, ignore=None)[source]
Read settings from a flat dictionary.
- fmriprep.config.load(filename, skip=None, init=True)[source]
Load settings from file.
- Parameters:
filename (
os.PathLike) – TOML file containing fMRIPrep configuration.skip (dict or None) – Sets of values to ignore during load, keyed by section name
init (bool or
Container) – Initialize all, none, or a subset of configurations.
Workflows
fMRIPrep base processing workflows
- fmriprep.workflows.base.init_fmriprep_wf()[source]
Build fMRIPrep’s pipeline.
This workflow organizes the execution of FMRIPREP, with a sub-workflow for each subject.
If FreeSurfer’s
recon-allis to be run, a corresponding folder is created and populated with any needed template subjects under the derivatives folder.- Workflow Graph
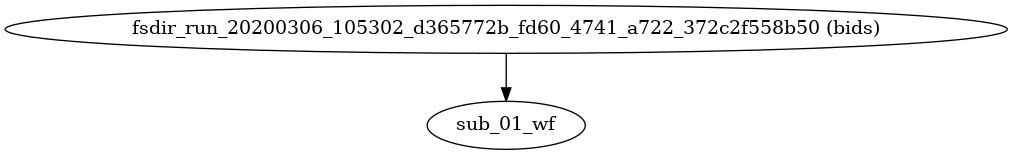
(Source code, png, svg, pdf)
- fmriprep.workflows.base.init_single_subject_wf(subject_id: str)[source]
Organize the preprocessing pipeline for a single subject.
It collects and reports information about the subject, and prepares sub-workflows to perform anatomical and functional preprocessing. Anatomical preprocessing is performed in a single workflow, regardless of the number of sessions. Functional preprocessing is performed using a separate workflow for each individual BOLD series.
- Workflow Graph
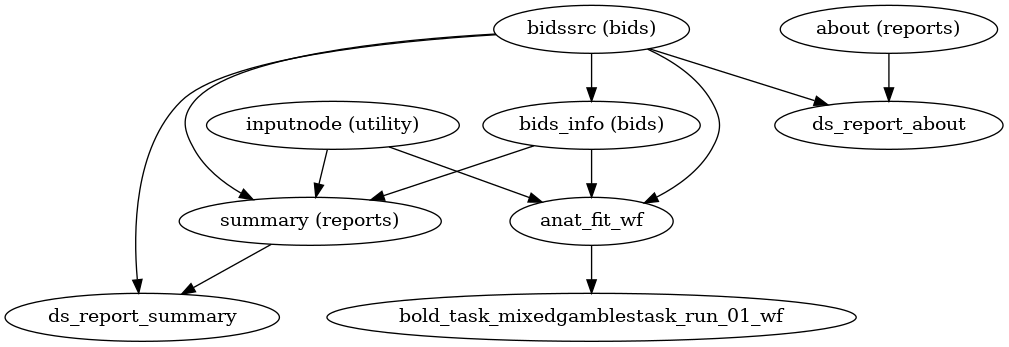
(Source code, png, svg, pdf)
Pre-processing fMRI - BOLD signal workflows
Orchestrating the BOLD-preprocessing workflow
- fmriprep.workflows.bold.base.init_bold_wf(*, bold_series: List[str], precomputed: dict = {}, fieldmap_id: str | None = None) Workflow[source]
This workflow controls the functional preprocessing stages of fMRIPrep.
- Workflow Graph
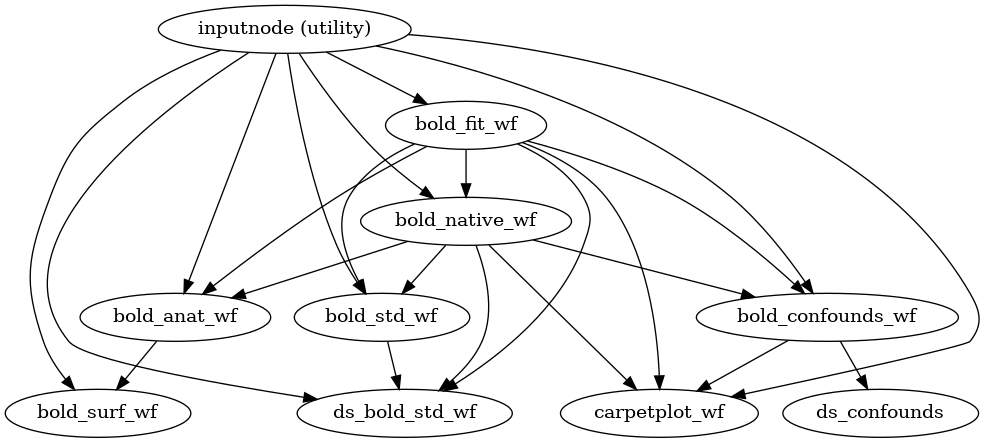
(Source code, png, svg, pdf)
- Parameters:
bold_series – List of paths to NIfTI files.
precomputed – Dictionary containing precomputed derivatives to reuse, if possible.
fieldmap_id – ID of the fieldmap to use to correct this BOLD series. If
None, no correction will be applied.
- Parameters:
t1w_preproc – Bias-corrected structural template image
t1w_mask – Mask of the skull-stripped template image
t1w_dseg – Segmentation of preprocessed structural image, including gray-matter (GM), white-matter (WM) and cerebrospinal fluid (CSF)
t1w_tpms – List of tissue probability maps in T1w space
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
fsnative2t1w_xfm – LTA-style affine matrix translating from FreeSurfer-conformed subject space to T1w
white – FreeSurfer white matter surfaces, in T1w space, collated left, then right
midthickness – FreeSurfer mid-thickness surfaces, in T1w space, collated left, then right
pial – FreeSurfer pial surfaces, in T1w space, collated left, then right
sphere_reg_fsLR – Registration spheres from fsnative to fsLR space, collated left, then right
anat_ribbon – Binary cortical ribbon mask in T1w space
fmap_id – Unique identifiers to select fieldmap files
fmap – List of estimated fieldmaps (collated with fmap_id)
fmap_ref – List of fieldmap reference files (collated with fmap_id)
fmap_coeff – List of lists of spline coefficient files (collated with fmap_id)
fmap_mask – List of fieldmap masks (collated with fmap_id)
sdc_method – List of fieldmap correction method names (collated with fmap_id)
anat2std_xfm – Transform from anatomical space to standard space
std_t1w – T1w reference image in standard space
std_mask – Brain (binary) mask of the standard reference image
std_space – Value of space entity to be used in standard space output filenames
std_resolution – Value of resolution entity to be used in standard space output filenames
std_cohort – Value of cohort entity to be used in standard space output filenames
anat2mni6_xfm – Transform from anatomical space to MNI152NLin6Asym space
mni6_mask – Brain (binary) mask of the MNI152NLin6Asym reference image
mni2009c2anat_xfm – Transform from MNI152NLin2009cAsym to anatomical space
Note that ``anat2std_xfm``, ``std_space``, ``std_resolution``,
``std_cohort``, ``std_t1w`` and ``std_mask`` are treated as single
inputs. In order to resample to multiple target spaces, connect
these fields to an iterable.
See also
init_bold_fit_wf()init_bold_native_wf()init_bold_volumetric_resample_wf()init_ds_bold_native_wf()init_ds_volumes_wf()init_t2s_reporting_wf()init_carpetplot_wf()
- fmriprep.workflows.bold.base.init_bold_fit_wf(*, bold_series: List[str], precomputed: dict = {}, fieldmap_id: str | None = None, omp_nthreads: int = 1, name: str = 'bold_fit_wf') Workflow[source]
This workflow controls the minimal estimation steps for functional preprocessing.
- Workflow Graph
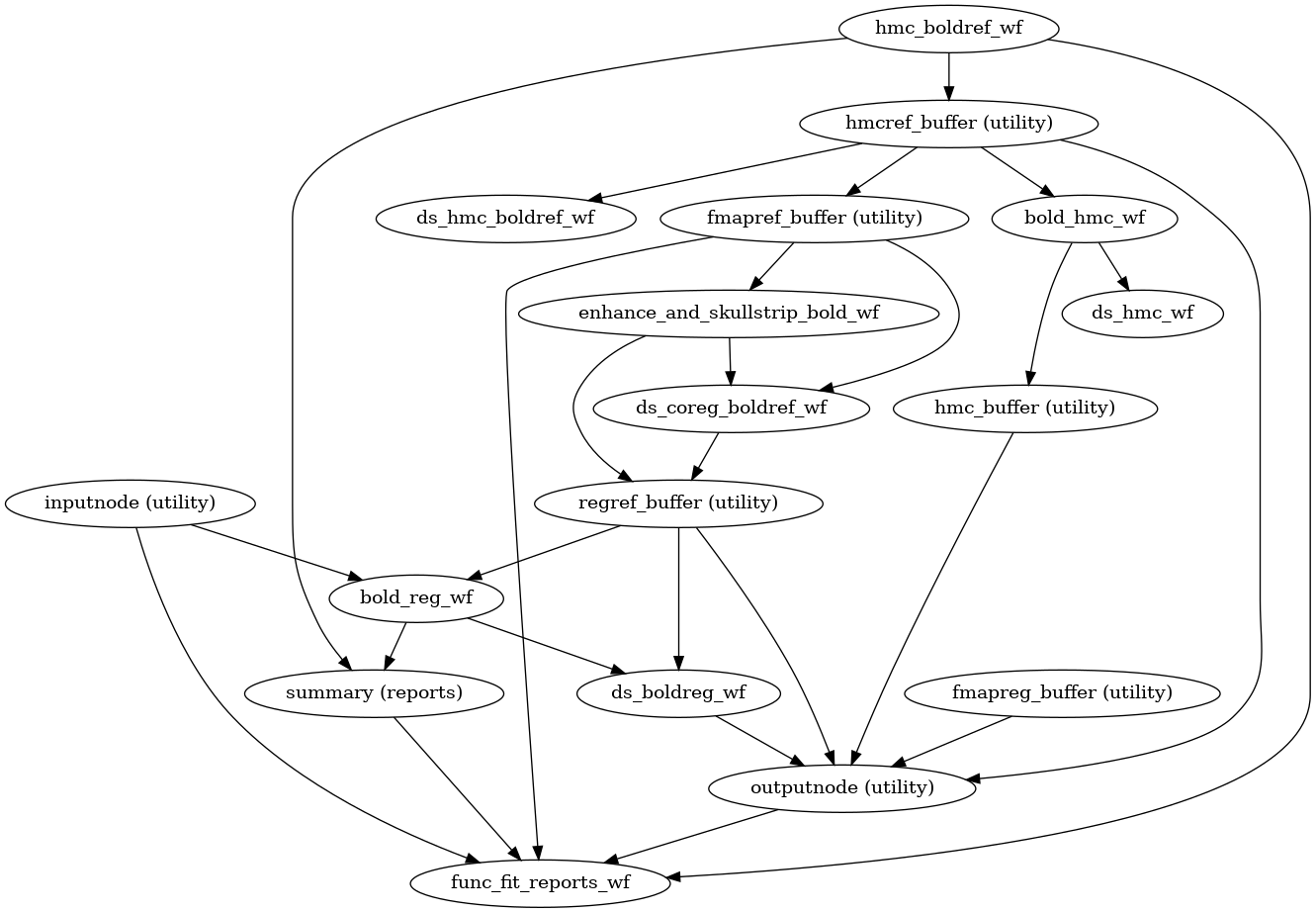
(Source code, png, svg, pdf)
- Parameters:
bold_series – List of paths to NIfTI files, sorted by echo time.
precomputed – Dictionary containing precomputed derivatives to reuse, if possible.
fieldmap_id – ID of the fieldmap to use to correct this BOLD series. If
None, no correction will be applied.
- Parameters:
bold_file – BOLD series NIfTI file
t1w_preproc – Bias-corrected structural template image
t1w_mask – Mask of the skull-stripped template image
t1w_dseg – Segmentation of preprocessed structural image, including gray-matter (GM), white-matter (WM) and cerebrospinal fluid (CSF)
anat2std_xfm – List of transform files, collated with templates
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
fsnative2t1w_xfm – LTA-style affine matrix translating from FreeSurfer-conformed subject space to T1w
fmap_id – Unique identifiers to select fieldmap files
fmap – List of estimated fieldmaps (collated with fmap_id)
fmap_ref – List of fieldmap reference files (collated with fmap_id)
fmap_coeff – List of lists of spline coefficient files (collated with fmap_id)
fmap_mask – List of fieldmap masks (collated with fmap_id)
sdc_method – List of fieldmap correction method names (collated with fmap_id)
- Parameters:
hmc_boldref – BOLD reference image used for head motion correction. Minimally processed to ensure consistent contrast with BOLD series.
coreg_boldref – BOLD reference image used for coregistration. Contrast-enhanced and fieldmap-corrected for greater anatomical fidelity, and aligned with
hmc_boldref.bold_mask – Mask of
coreg_boldref.motion_xfm – Affine transforms from each BOLD volume to
hmc_boldref, written as concatenated ITK affine transforms.boldref2anat_xfm – Affine transform mapping from BOLD reference space to the anatomical space.
boldref2fmap_xfm – Affine transform mapping from BOLD reference space to the fieldmap space, if applicable.
movpar_file – MCFLIRT motion parameters, normalized to SPM format (X, Y, Z, Rx, Ry, Rz)
rmsd_file – Root mean squared deviation as measured by
fsl_motion_outliers[Jenkinson2002].dummy_scans – The number of dummy scans declared or detected at the beginning of the series.
See also
init_raw_boldref_wf()init_enhance_and_skullstrip_bold_wf()init_ds_boldref_wf()init_ds_hmc_wf()init_ds_registration_wf()
- fmriprep.workflows.bold.base.init_bold_native_wf(*, bold_series: List[str], fieldmap_id: str | None = None, omp_nthreads: int = 1, name: str = 'bold_native_wf') Workflow[source]
Minimal resampling workflow.
This workflow performs slice-timing correction, and resamples to boldref space with head motion and susceptibility distortion correction. It also handles multi-echo processing and selects the transforms needed to perform further resampling.
- Workflow Graph
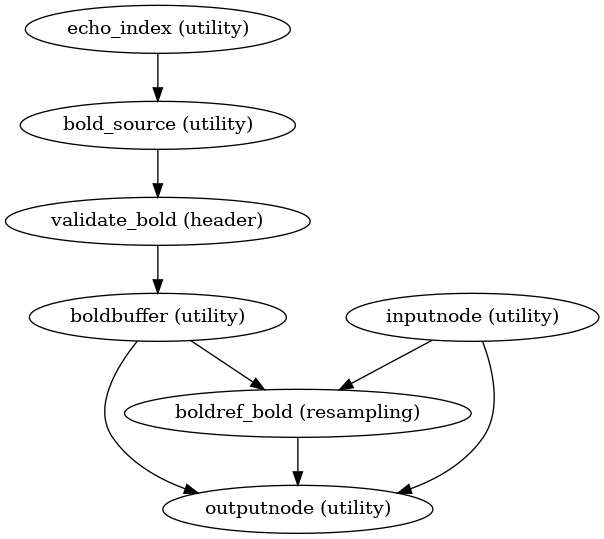
(Source code, png, svg, pdf)
- Parameters:
bold_series – List of paths to NIfTI files.
fieldmap_id – ID of the fieldmap to use to correct this BOLD series. If
None, no correction will be applied.
- Parameters:
boldref – BOLD reference file
bold_mask – Mask of BOLD reference file
motion_xfm – Affine transforms from each BOLD volume to
hmc_boldref, written as concatenated ITK affine transforms.boldref2fmap_xfm – Affine transform mapping from BOLD reference space to the fieldmap space, if applicable.
fmap_id – Unique identifiers to select fieldmap files
fmap_ref – List of fieldmap reference files (collated with fmap_id)
fmap_coeff – List of lists of spline coefficient files (collated with fmap_id)
- Parameters:
bold_minimal – BOLD series ready for further resampling. For single-echo data, only slice-timing correction (STC) may have been applied. For multi-echo data, this is identical to bold_native.
bold_native – BOLD series resampled into BOLD reference space. Slice-timing, head motion and susceptibility distortion correction (STC, HMC, SDC) will all be applied to each file. For multi-echo data, the echos are combined to form an optimal combination.
metadata – Metadata dictionary of BOLD series with the shortest echo
motion_xfm – Motion correction transforms for further correcting bold_minimal. For multi-echo data, motion correction has already been applied, so this will be undefined.
bold_echos – The individual, corrected echos, suitable for use in Tedana. (Multi-echo only.)
t2star_map – The T2* map estimated by Tedana when calculating the optimal combination. (Multi-echo only.)
See also
Head-Motion Estimation and Correction (HMC) of BOLD images
- fmriprep.workflows.bold.hmc.init_bold_hmc_wf(mem_gb: float, omp_nthreads: int, name: str = 'bold_hmc_wf')[source]
Build a workflow to estimate head-motion parameters.
This workflow estimates the motion parameters to perform HMC over the input BOLD image.
- Workflow Graph
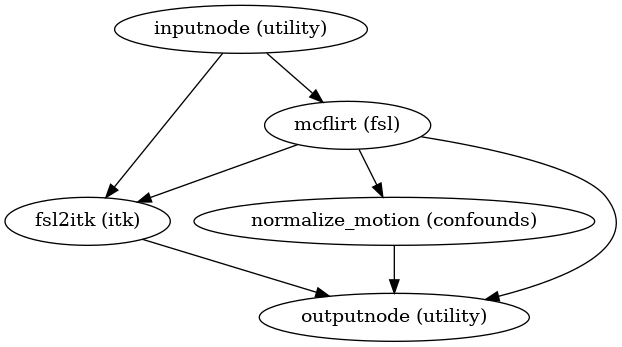
(Source code, png, svg, pdf)
- Parameters:
- Parameters:
bold_file – BOLD series NIfTI file
raw_ref_image – Reference image to which BOLD series is motion corrected
- Parameters:
xforms – ITKTransform file aligning each volume to
ref_imagemovpar_file – MCFLIRT motion parameters, normalized to SPM format (X, Y, Z, Rx, Ry, Rz)
rmsd_file – Root mean squared deviation as measured by
fsl_motion_outliers[Jenkinson2002].
Slice-Timing Correction (STC) of BOLD images
- fmriprep.workflows.bold.stc.init_bold_stc_wf(*, mem_gb: dict, metadata: dict, name='bold_stc_wf')[source]
Create a workflow for STC.
This workflow performs STC over the input BOLD image.
- Workflow Graph
- Parameters:
- Parameters:
bold_file – BOLD series NIfTI file
skip_vols – Number of non-steady-state volumes detected at beginning of
bold_file
- Parameters:
stc_file – Slice-timing corrected BOLD series NIfTI file
Generate T2* map from multi-echo BOLD images
- fmriprep.workflows.bold.t2s.init_bold_t2s_wf(echo_times: Sequence[float], mem_gb: float, omp_nthreads: int, name: str = 'bold_t2s_wf')[source]
Combine multiple echos of ME-EPI.
This workflow wraps the tedana T2* workflow to optimally combine multiple preprocessed echos and derive a T2★ map. The following steps are performed: #. Compute the T2★ map #. Create an optimally combined ME-EPI time series
- Parameters:
- Parameters:
bold_file – list of individual echo files
bold_mask – a binary mask to apply to the BOLD files
- Parameters:
bold – the optimally combined time series for all supplied echos
t2star_map – the calculated T2★ map
Registration workflows
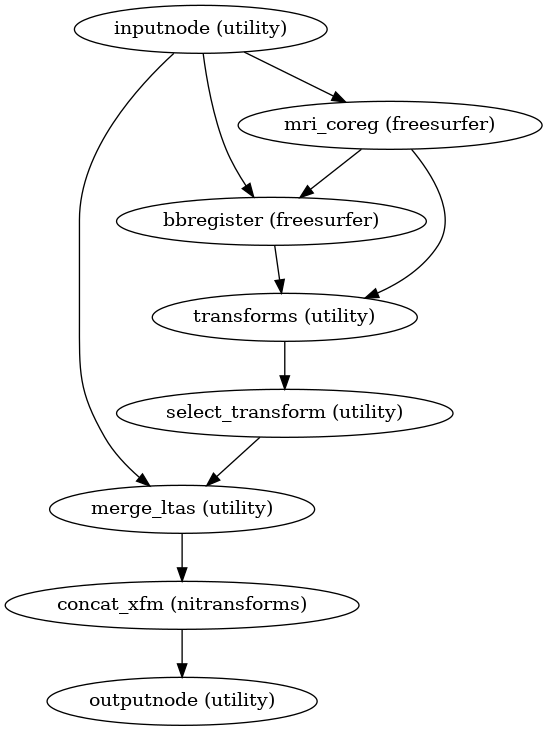
- fmriprep.workflows.bold.registration.init_bold_reg_wf(freesurfer: bool, use_bbr: bool, bold2t1w_dof: Literal[6, 9, 12], bold2t1w_init: Literal['register', 'header'], mem_gb: float, omp_nthreads: int, name: str = 'bold_reg_wf', sloppy: bool = False)[source]
Build a workflow to run same-subject, BOLD-to-T1w image-registration.
Calculates the registration between a reference BOLD image and T1w-space using a boundary-based registration (BBR) cost function. If FreeSurfer-based preprocessing is enabled, the
bbregisterutility is used to align the BOLD images to the reconstructed subject, and the resulting transform is adjusted to target the T1 space. If FreeSurfer-based preprocessing is disabled, FSL FLIRT is used with the BBR cost function to directly target the T1 space.- Workflow Graph
(Source code, png, svg, pdf)
- Parameters:
freesurfer (
bool) – Enable FreeSurfer functional registration (bbregister)use_bbr (
boolor None) – Enable/disable boundary-based registration refinement. IfNone, test BBR result for distortion before accepting.bold2t1w_dof (6, 9 or 12) – Degrees-of-freedom for BOLD-T1w registration
bold2t1w_init (str, ‘header’ or ‘register’) – If
'header', use header information for initialization of BOLD and T1 images. If'register', align volumes by their centers.mem_gb (
float) – Size of BOLD file in GBomp_nthreads (
int) – Maximum number of threads an individual process may usename (
str) – Name of workflow (default:bold_reg_wf)
- Parameters:
ref_bold_brain – Reference image to which BOLD series is aligned If
fieldwarp == True,ref_bold_brainshould be unwarpedt1w_brain – Skull-stripped
t1w_preproct1w_dseg – Segmentation of preprocessed structural image, including gray-matter (GM), white-matter (WM) and cerebrospinal fluid (CSF)
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
fsnative2t1w_xfm – LTA-style affine matrix translating from FreeSurfer-conformed subject space to T1w
- Parameters:
itk_bold_to_t1 – Affine transform from
ref_bold_brainto T1 space (ITK format)itk_t1_to_bold – Affine transform from T1 space to BOLD space (ITK format)
fallback – Boolean indicating whether BBR was rejected (mri_coreg registration returned)
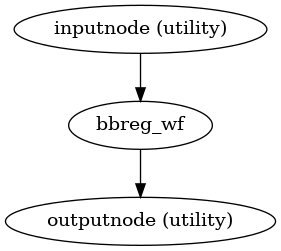
- fmriprep.workflows.bold.registration.init_bbreg_wf(use_bbr: bool, bold2t1w_dof: Literal[6, 9, 12], bold2t1w_init: Literal['register', 'header'], omp_nthreads: int, name: str = 'bbreg_wf')[source]
Build a workflow to run FreeSurfer’s
bbregister.This workflow uses FreeSurfer’s
bbregisterto register a BOLD image to a T1-weighted structural image.It is a counterpart to
init_fsl_bbr_wf(), which performs the same task using FSL’s FLIRT with a BBR cost function. Theuse_bbroption permits a high degree of control over registration. IfFalse, standard, affine coregistration will be performed using FreeSurfer’smri_coregtool. IfTrue,bbregisterwill be seeded with the initial transform found bymri_coreg(equivalent to runningbbregister --init-coreg). IfNone, afterbbregisteris run, the resulting affine transform will be compared to the initial transform found bymri_coreg. Excessive deviation will result in rejecting the BBR refinement and accepting the original, affine registration.- Workflow Graph
(Source code, png, svg, pdf)
- Parameters:
use_bbr (
boolor None) – Enable/disable boundary-based registration refinement. IfNone, test BBR result for distortion before accepting.bold2t1w_dof (6, 9 or 12) – Degrees-of-freedom for BOLD-T1w registration
bold2t1w_init (str, ‘header’ or ‘register’) – If
'header', use header information for initialization of BOLD and T1 images. If'register', align volumes by their centers.name (
str, optional) – Workflow name (default: bbreg_wf)
- Parameters:
in_file – Reference BOLD image to be registered
fsnative2t1w_xfm – FSL-style affine matrix translating from FreeSurfer T1.mgz to T1w
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID (must have folder in SUBJECTS_DIR)
t1w_preproc – Unused (see
init_fsl_bbr_wf())t1w_mask – Unused (see
init_fsl_bbr_wf())t1w_dseg – Unused (see
init_fsl_bbr_wf())
- Parameters:
itk_bold_to_t1 – Affine transform from
ref_bold_brainto T1 space (ITK format)itk_t1_to_bold – Affine transform from T1 space to BOLD space (ITK format)
fallback – Boolean indicating whether BBR was rejected (mri_coreg registration returned)
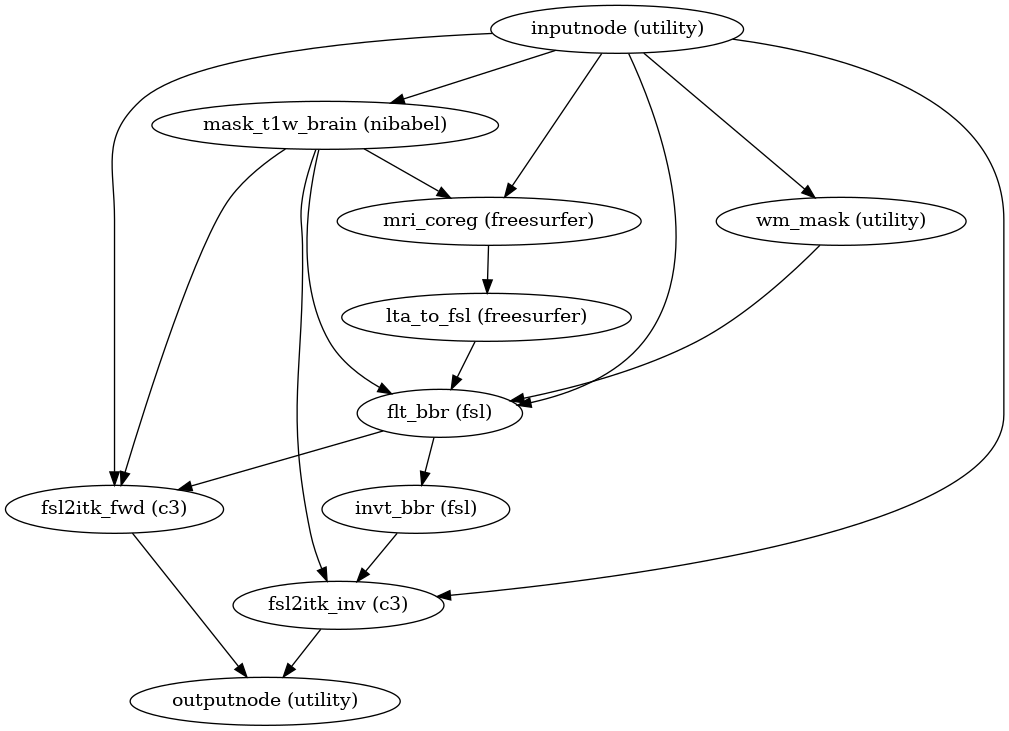
- fmriprep.workflows.bold.registration.init_fsl_bbr_wf(use_bbr: bool, bold2t1w_dof: Literal[6, 9, 12], bold2t1w_init: Literal['register', 'header'], omp_nthreads: int, sloppy: bool = False, name: str = 'fsl_bbr_wf')[source]
Build a workflow to run FSL’s
flirt.This workflow uses FSL FLIRT to register a BOLD image to a T1-weighted structural image, using a boundary-based registration (BBR) cost function. It is a counterpart to
init_bbreg_wf(), which performs the same task using FreeSurfer’sbbregister.The
use_bbroption permits a high degree of control over registration. IfFalse, standard, rigid coregistration will be performed by FLIRT. IfTrue, FLIRT-BBR will be seeded with the initial transform found by the rigid coregistration. IfNone, after FLIRT-BBR is run, the resulting affine transform will be compared to the initial transform found by FLIRT. Excessive deviation will result in rejecting the BBR refinement and accepting the original, affine registration.- Workflow Graph
(Source code, png, svg, pdf)
- Parameters:
use_bbr (
boolor None) – Enable/disable boundary-based registration refinement. IfNone, test BBR result for distortion before accepting.bold2t1w_dof (6, 9 or 12) – Degrees-of-freedom for BOLD-T1w registration
bold2t1w_init (str, ‘header’ or ‘register’) – If
'header', use header information for initialization of BOLD and T1 images. If'register', align volumes by their centers.name (
str, optional) – Workflow name (default: fsl_bbr_wf)
- Parameters:
in_file – Reference BOLD image to be registered
t1w_preproc – T1-weighted structural image
t1w_mask – Brain mask of structural image
t1w_dseg – FAST segmentation of masked
t1w_preprocfsnative2t1w_xfm – Unused (see
init_bbreg_wf())subjects_dir – Unused (see
init_bbreg_wf())subject_id – Unused (see
init_bbreg_wf())
- Parameters:
itk_bold_to_t1 – Affine transform from
ref_bold_brainto T1w space (ITK format)itk_t1_to_bold – Affine transform from T1 space to BOLD space (ITK format)
fallback – Boolean indicating whether BBR was rejected (rigid FLIRT registration returned)
Resampling workflows
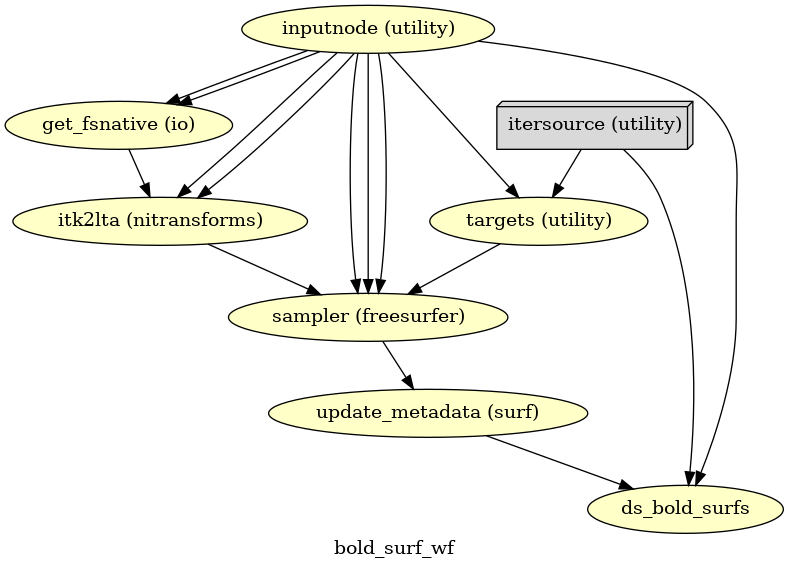
- fmriprep.workflows.bold.resampling.init_bold_surf_wf(*, mem_gb: float, surface_spaces: List[str], medial_surface_nan: bool, metadata: dict, output_dir: str, name: str = 'bold_surf_wf')[source]
Sample functional images to FreeSurfer surfaces.
For each vertex, the cortical ribbon is sampled at six points (spaced 20% of thickness apart) and averaged.
Outputs are in GIFTI format.
- Workflow Graph
(Source code, png, svg, pdf)
- Parameters:
surface_spaces (
list) – List of FreeSurfer surface-spaces (eitherfsaverage{3,4,5,6,}orfsnative) the functional images are to be resampled to. Forfsnative, images will be resampled to the individual subject’s native surface.medial_surface_nan (
bool) – Replace medial wall values with NaNs on functional GIFTI files
- Parameters:
source_file – Original BOLD series
bold_t1w – Motion-corrected BOLD series in T1 space
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
fsnative2t1w_xfm – ITK-style affine matrix translating from FreeSurfer-conformed subject space to T1w
- Parameters:
surfaces – BOLD series, resampled to FreeSurfer surfaces
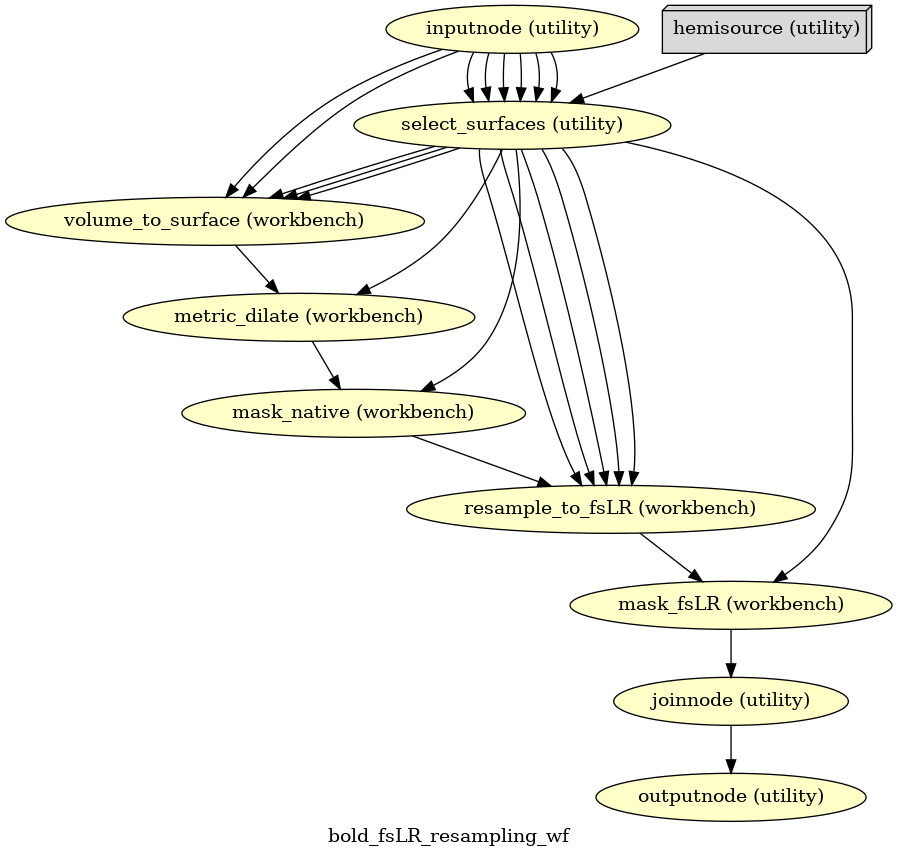
- fmriprep.workflows.bold.resampling.init_bold_fsLR_resampling_wf(grayord_density: Literal['91k', '170k'], omp_nthreads: int, mem_gb: float, name: str = 'bold_fsLR_resampling_wf')[source]
Resample BOLD time series to fsLR surface.
This workflow is derived heavily from three scripts within the DCAN-HCP pipelines scripts
Line numbers correspond to the locations of the code in the original scripts, found at: https://github.com/DCAN-Labs/DCAN-HCP/tree/9291324/
- Workflow Graph
(Source code, png, svg, pdf)
- Parameters:
- Parameters:
bold_file (
str) – Path to BOLD file resampled into T1 spacewhite (
listofstr) – Path to left and right hemisphere white matter GIFTI surfaces.pial (
listofstr) – Path to left and right hemisphere pial GIFTI surfaces.midthickness (
listofstr) – Path to left and right hemisphere midthickness GIFTI surfaces.midthickness_fsLR (
listofstr) – Path to left and right hemisphere midthickness GIFTI surfaces in fsLR space.sphere_reg_fsLR (
listofstr) – Path to left and right hemisphere sphere.reg GIFTI surfaces, mapping from subject to fsLRcortex_mask (
listofstr) – Path to left and right hemisphere cortical masks.volume_roi (
stror Undefined) – Pre-calculated goodvoxels mask. Not required.
- Parameters:
bold_fsLR (
listofstr) – Path to BOLD series resampled as functional GIFTI files in fsLR space
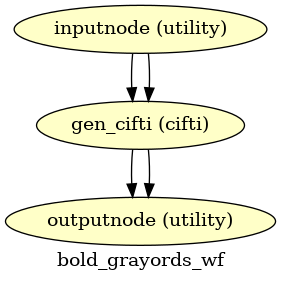
- fmriprep.workflows.bold.resampling.init_bold_grayords_wf(grayord_density: Literal['91k', '170k'], mem_gb: float, repetition_time: float, name: str = 'bold_grayords_wf')[source]
Sample Grayordinates files onto the fsLR atlas.
Outputs are in CIFTI2 format.
- Workflow Graph
(Source code, png, svg, pdf)
- Parameters:
- Parameters:
- Parameters:
- fmriprep.workflows.bold.resampling.init_goodvoxels_bold_mask_wf(mem_gb: float, name: str = 'goodvoxels_bold_mask_wf')[source]
Calculate a mask of a BOLD series excluding high variance voxels.
- Workflow Graph
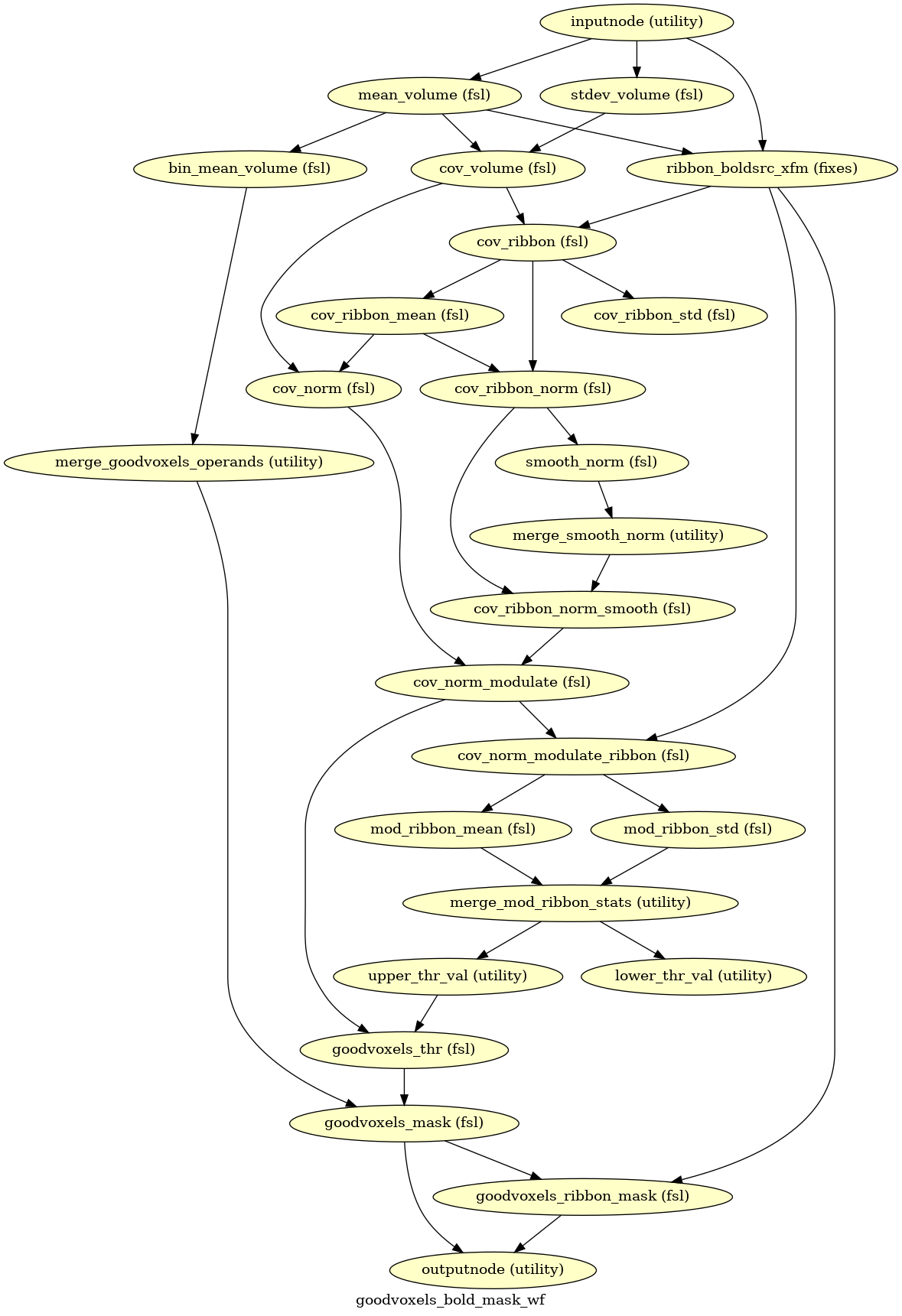
(Source code, png, svg, pdf)
- Parameters:
- Parameters:
anat_ribbon – Cortical ribbon in T1w space
bold_file – Motion-corrected BOLD series in T1w space
- Parameters:
masked_bold – BOLD series after masking outlier voxels with locally high COV
goodvoxels_ribbon – Cortical ribbon mask excluding voxels with locally high COV
Calculate BOLD confounds
- fmriprep.workflows.bold.confounds.init_bold_confs_wf(mem_gb: float, metadata: dict, regressors_all_comps: bool, regressors_dvars_th: float, regressors_fd_th: float, freesurfer: bool = False, name: str = 'bold_confs_wf')[source]
Build a workflow to generate and write out confounding signals.
This workflow calculates confounds for a BOLD series, and aggregates them into a TSV file, for use as nuisance regressors in a GLM. The following confounds are calculated, with column headings in parentheses:
Region-wise average signal (
csf,white_matter,global_signal)DVARS - original and standardized variants (
dvars,std_dvars)Framewise displacement, based on head-motion parameters (
framewise_displacement)Temporal CompCor (
t_comp_cor_XX)Anatomical CompCor (
a_comp_cor_XX)Cosine basis set for high-pass filtering w/ 0.008 Hz cut-off (
cosine_XX)Non-steady-state volumes (
non_steady_state_XX)Estimated head-motion parameters, in mm and rad (
trans_x,trans_y,trans_z,rot_x,rot_y,rot_z)
Prior to estimating aCompCor and tCompCor, non-steady-state volumes are censored and high-pass filtered using a DCT basis. The cosine basis, as well as one regressor per censored volume, are included for convenience.
- Workflow Graph
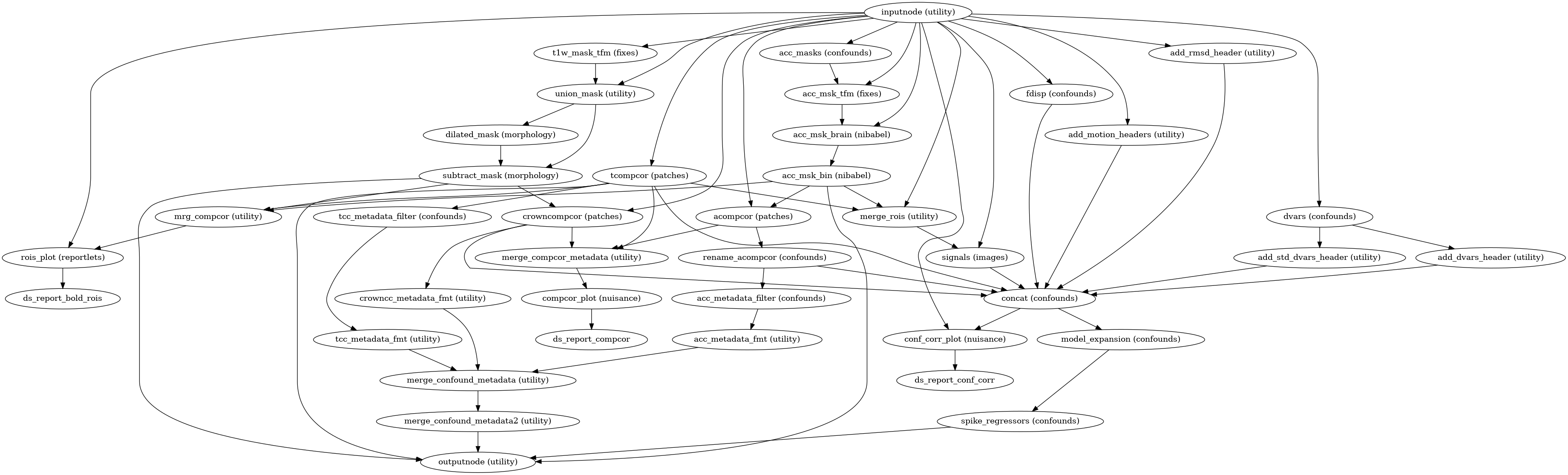
(Source code, png, svg, pdf)
- Parameters:
mem_gb (
float) – Size of BOLD file in GB - please note that this size should be calculated after resamplings that may extend the FoVmetadata (
dict) – BIDS metadata for BOLD filename (
str) – Name of workflow (default:bold_confs_wf)regressors_all_comps (
bool) – Indicates whether CompCor decompositions should return all components instead of the minimal number of components necessary to explain 50 percent of the variance in the decomposition mask.regressors_dvars_th (
float) – Criterion for flagging DVARS outliersregressors_fd_th (
float) – Criterion for flagging framewise displacement outliers
- Parameters:
bold – BOLD image, after the prescribed corrections (STC, HMC and SDC) when available.
bold_mask – BOLD series mask
movpar_file – SPM-formatted motion parameters file
rmsd_file – Root mean squared deviation as measured by
fsl_motion_outliers[Jenkinson2002].skip_vols – number of non steady state volumes
t1w_mask – Mask of the skull-stripped template image
t1w_tpms – List of tissue probability maps in T1w space
boldref2anat_xfm – Affine matrix that maps the BOLD reference space into alignment with the anatomical (T1w) space
- Parameters:
confounds_file – TSV of all aggregated confounds
rois_report – Reportlet visualizing white-matter/CSF mask used for aCompCor, the ROI for tCompCor and the BOLD brain mask.
confounds_metadata – Confounds metadata dictionary.
crown_mask – Mask of brain edge voxels